Os09g0459600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Calmodulin-binding protein phosphatase.
FiT-DB / Search/ Help/ Sample detail
|
Os09g0459600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Calmodulin-binding protein phosphatase.
|
|
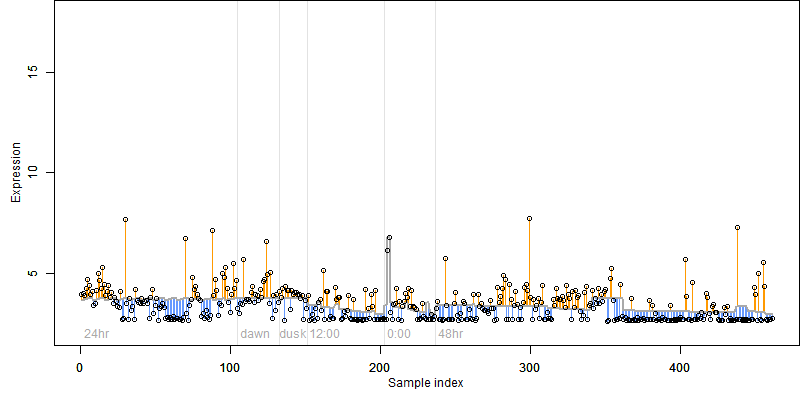
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
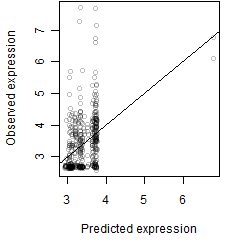
Dependence on each variable
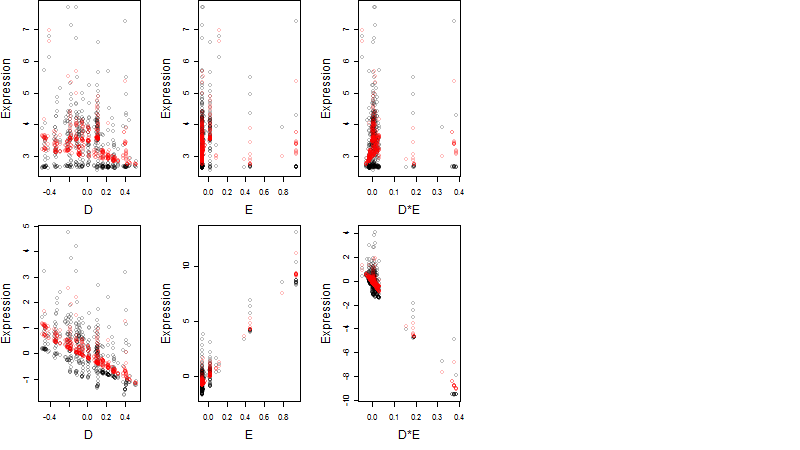
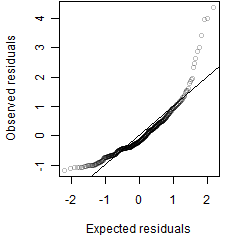
Residual plot

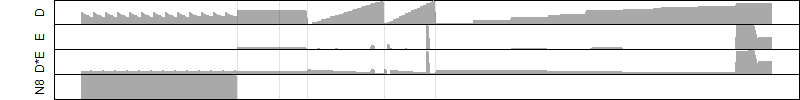
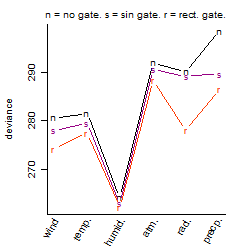
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 264.14 | 0.764 | 3.77 | -1.91 | 0.237 | 9.78 | -0.911 | -23.5 | 0.477 | 6.13 | 50 | 4323 | -- | -- |
| 304.31 | 0.819 | 3.33 | -0.45 | 0.274 | 0.226 | -0.989 | -- | 0.317 | 4.8 | 50 | 4323 | -- | -- |
| 266.01 | 0.76 | 3.77 | -1.92 | 0.237 | 9.79 | -- | -23.5 | 0.478 | 6.96 | 50 | 4332 | -- | -- |
| 287.95 | 0.79 | 3.3 | -0.61 | 0.299 | 1.14 | -- | -- | 0.478 | 5.66 | 58.5 | 1844 | -- | -- |
| 268.98 | 0.764 | 3.77 | -1.91 | -- | 9.8 | -- | -23.5 | 0.487 | -- | 50.01 | 4315 | -- | -- |
| 304.47 | 0.818 | 3.33 | -0.439 | 0.27 | -- | -1.01 | -- | 0.316 | 4.79 | -- | -- | -- | -- |
| 307.13 | 0.821 | 3.33 | -0.435 | 0.264 | -- | -- | -- | 0.321 | 4.92 | -- | -- | -- | -- |
| 291.75 | 0.796 | 3.3 | -0.631 | -- | 1.21 | -- | -- | 0.472 | -- | 57.51 | 1831 | -- | -- |
| 298.29 | 0.804 | 3.28 | -- | 0.273 | 0.878 | -- | -- | 0.519 | 5.8 | 60 | 1844 | -- | -- |
| 310.8 | 0.824 | 3.33 | -0.425 | -- | -- | -- | -- | 0.328 | -- | -- | -- | -- | -- |
| 312.11 | 0.826 | 3.32 | -- | 0.256 | -- | -- | -- | 0.369 | 4.97 | -- | -- | -- | -- |
| 302.02 | 0.809 | 3.29 | -- | -- | 0.912 | -- | -- | 0.519 | -- | 59 | 1824 | -- | -- |
| 315.57 | 0.829 | 3.32 | -- | -- | -- | -- | -- | 0.375 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance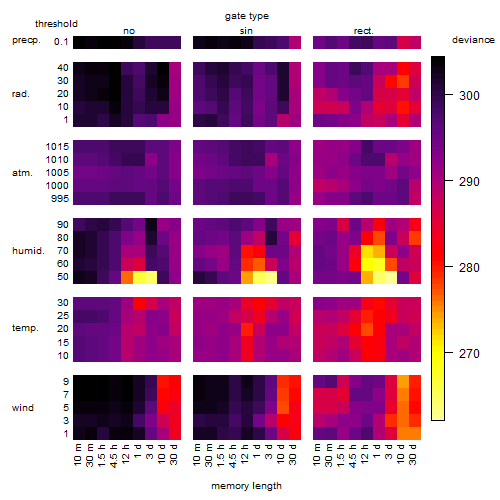
|
Histogram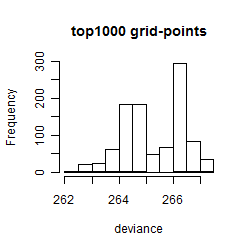 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 262.11 | humidity | 50 | 4320 | < th | dose dependent | rect. | 14 | 3 |
| 22 | 262.92 | humidity | 50 | 4320 | < th | dose dependent | sin | 14 | NA |
| 26 | 263.12 | humidity | 50 | 4320 | < th | dose independent | rect. | 17 | 23 |
| 69 | 263.72 | humidity | 50 | 4320 | < th | dose independent | sin | 1 | NA |
| 90 | 263.88 | humidity | 50 | 4320 | < th | dose independent | rect. | 12 | 23 |
| 93 | 263.90 | humidity | 50 | 4320 | < th | dose independent | rect. | 12 | 6 |
| 112 | 264.06 | humidity | 50 | 4320 | < th | dose independent | rect. | 13 | 3 |
| 131 | 264.14 | humidity | 50 | 4320 | < th | dose dependent | no | NA | NA |
| 308 | 264.52 | humidity | 50 | 4320 | < th | dose independent | no | NA | NA |
| 683 | 266.33 | humidity | 50 | 4320 | > th | dose independent | rect. | 17 | 14 |
| 822 | 266.36 | humidity | 50 | 4320 | > th | dose independent | rect. | 17 | 16 |
| 858 | 266.49 | humidity | 50 | 4320 | > th | dose independent | rect. | 17 | 11 |
| 882 | 266.51 | humidity | 50 | 4320 | > th | dose independent | rect. | 17 | 5 |
| 886 | 266.69 | humidity | 50 | 4320 | > th | dose independent | rect. | 17 | 1 |
| 894 | 266.84 | humidity | 50 | 4320 | > th | dose independent | rect. | 17 | 8 |
| 994 | 267.15 | humidity | 50 | 1440 | < th | dose independent | rect. | 17 | 21 |