Os09g0323900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Haem peroxidase family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os09g0323900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Haem peroxidase family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
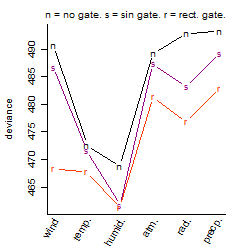
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 467.05 | 1.02 | 3.23 | 0.799 | 0.112 | -7.14 | 0.942 | 26.8 | -0.271 | 13.6 | 49.67 | 297 | -- | -- |
| 504.85 | 1.05 | 3.32 | 0.582 | 0.16 | -0.309 | 1.38 | -- | -0.256 | 14 | 49.67 | 297 | -- | -- |
| 470.35 | 1.01 | 3.22 | 0.788 | 0.113 | -7.48 | -- | 28 | -0.266 | 13.2 | 49.41 | 306 | -- | -- |
| 479.98 | 1.02 | 3.35 | 0.555 | 0.152 | 3.49 | -- | -- | -0.267 | 9.54 | 48 | 353 | -- | -- |
| 471.03 | 1.01 | 3.22 | 0.793 | -- | -7.56 | -- | 28.2 | -0.268 | -- | 49.39 | 305 | -- | -- |
| 505.13 | 1.05 | 3.32 | 0.569 | 0.151 | -- | 1.34 | -- | -0.255 | 14 | -- | -- | -- | -- |
| 511.8 | 1.06 | 3.31 | 0.608 | 0.158 | -- | -- | -- | -0.248 | 12.7 | -- | -- | -- | -- |
| 477.66 | 1.02 | 3.31 | 0.399 | -- | 4.06 | -- | -- | -0.228 | -- | 47 | 295 | -- | -- |
| 480.83 | 1.02 | 3.32 | -- | 0.128 | 4.28 | -- | -- | -0.27 | 9.55 | 46.73 | 296 | -- | -- |
| 513.37 | 1.06 | 3.32 | 0.603 | -- | -- | -- | -- | -0.25 | -- | -- | -- | -- | -- |
| 521.56 | 1.07 | 3.33 | -- | 0.149 | -- | -- | -- | -0.315 | 12.5 | -- | -- | -- | -- |
| 481.82 | 1.02 | 3.32 | -- | -- | 4.32 | -- | -- | -0.276 | -- | 47 | 295 | -- | -- |
| 522.95 | 1.07 | 3.33 | -- | -- | -- | -- | -- | -0.317 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 461.34 | humidity | 50 | 270 | < th | dose independent | rect. | 14 | 2 |
| 2 | 461.44 | humidity | 50 | 270 | < th | dose dependent | rect. | 14 | 2 |
| 4 | 461.73 | humidity | 50 | 270 | > th | dose independent | sin | 13 | NA |
| 22 | 461.85 | humidity | 50 | 270 | < th | dose dependent | sin | 12 | NA |
| 25 | 461.92 | humidity | 50 | 270 | < th | dose independent | sin | 12 | NA |
| 206 | 467.77 | temperature | 30 | 270 | > th | dose independent | rect. | 15 | 1 |
| 243 | 468.33 | wind | 1 | 10 | < th | dose independent | rect. | 14 | 4 |
| 282 | 468.76 | humidity | 50 | 270 | < th | dose dependent | no | NA | NA |
| 453 | 470.33 | temperature | 30 | 270 | > th | dose independent | rect. | 14 | 21 |
| 479 | 470.61 | wind | 1 | 90 | < th | dose dependent | rect. | 14 | 1 |
| 503 | 471.15 | wind | 3 | 90 | < th | dose independent | rect. | 15 | 4 |
| 507 | 471.55 | wind | 3 | 270 | < th | dose independent | rect. | 15 | 1 |
| 540 | 471.71 | temperature | 30 | 270 | > th | dose independent | sin | 18 | NA |
| 561 | 471.82 | humidity | 50 | 270 | < th | dose independent | no | NA | NA |
| 682 | 471.86 | wind | 3 | 90 | < th | dose dependent | rect. | 15 | 4 |
| 697 | 472.49 | wind | 1 | 10 | < th | dose dependent | rect. | 14 | 4 |
| 703 | 472.58 | temperature | 30 | 270 | < th | dose independent | no | NA | NA |
| 706 | 472.71 | wind | 3 | 10 | < th | dose independent | rect. | 16 | 1 |
| 709 | 472.86 | temperature | 30 | 270 | > th | dose independent | rect. | 10 | 9 |
| 724 | 472.90 | temperature | 30 | 270 | < th | dose independent | sin | 23 | NA |
| 725 | 472.97 | humidity | 50 | 270 | < th | dose dependent | sin | 6 | NA |
| 784 | 473.40 | temperature | 30 | 270 | > th | dose independent | no | NA | NA |
| 916 | 473.86 | humidity | 50 | 270 | > th | dose independent | no | NA | NA |
| 922 | 474.04 | temperature | 30 | 270 | < th | dose independent | rect. | 2 | 23 |
| 927 | 474.30 | temperature | 30 | 270 | < th | dose independent | sin | 14 | NA |
| 954 | 475.10 | wind | 5 | 90 | > th | dose dependent | rect. | 8 | 1 |
| 980 | 475.31 | wind | 3 | 10 | < th | dose dependent | rect. | 16 | 1 |