Os09g0260200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.

FiT-DB / Search/ Help/ Sample detail
|
Os09g0260200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
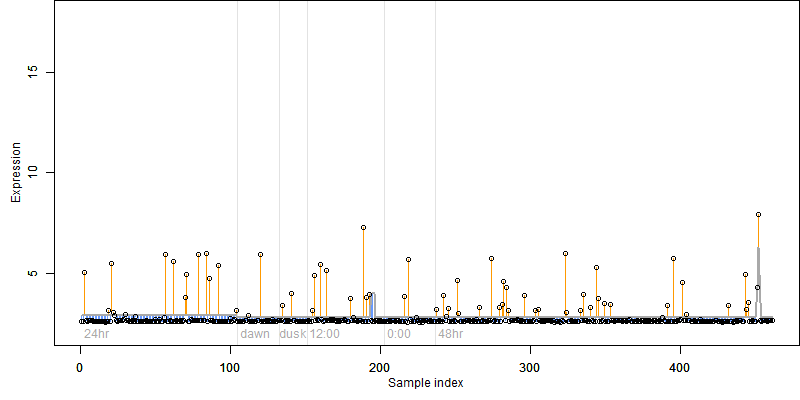
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

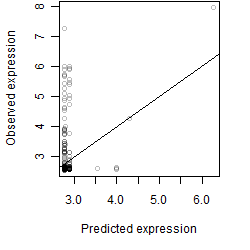
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 206.25 | 0.675 | 2.77 | -0.0765 | 0.0922 | -2.48 | 0.223 | 15.2 | 0.073 | 1.05 | 49 | 177 | -- | -- |
| 207.19 | 0.67 | 2.82 | -0.281 | 0.114 | 3.8 | -0.0641 | -- | 0.0336 | 2.51 | 47 | 158 | -- | -- |
| 205.87 | 0.668 | 2.78 | -0.117 | 0.108 | -2.44 | -- | 15.1 | 0.0712 | 2.93 | 47.14 | 158 | -- | -- |
| 206.94 | 0.67 | 2.81 | -0.236 | 0.112 | 3.74 | -- | -- | 0.0738 | 2.95 | 47.35 | 158 | -- | -- |
| 205.76 | 0.668 | 2.72 | -0.00226 | -- | -10.3 | -- | 33.4 | 0.0602 | -- | 47.8 | 157 | -- | -- |
| 226.48 | 0.706 | 2.81 | -0.0779 | 0.0305 | -- | -0.772 | -- | 0.0612 | 5.82 | -- | -- | -- | -- |
| 227.91 | 0.707 | 2.81 | -0.084 | 0.035 | -- | -- | -- | 0.0639 | 3.01 | -- | -- | -- | -- |
| 207.65 | 0.671 | 2.81 | -0.225 | -- | 3.7 | -- | -- | 0.0807 | -- | 47 | 158 | -- | -- |
| 208.3 | 0.672 | 2.8 | -- | 0.103 | 3.56 | -- | -- | 0.0973 | 2.78 | 47 | 158 | -- | -- |
| 227.98 | 0.706 | 2.81 | -0.0825 | -- | -- | -- | -- | 0.0648 | -- | -- | -- | -- | -- |
| 228.09 | 0.706 | 2.81 | -- | 0.0334 | -- | -- | -- | 0.0733 | 3 | -- | -- | -- | -- |
| 208.92 | 0.673 | 2.8 | -- | -- | 3.49 | -- | -- | 0.101 | -- | 47 | 158 | -- | -- |
| 228.16 | 0.705 | 2.81 | -- | -- | -- | -- | -- | 0.074 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance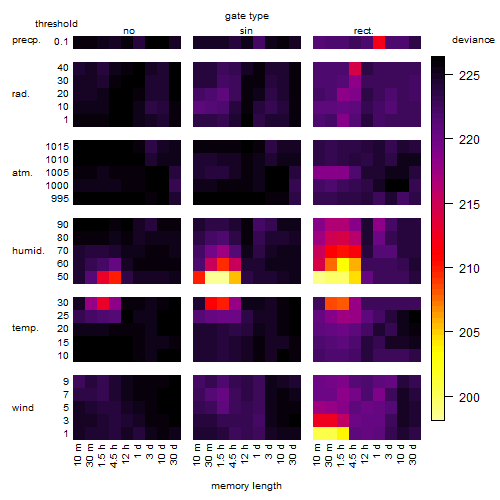
|
Histogram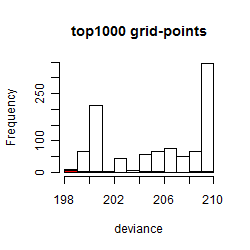 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 198.12 | humidity | 50 | 30 | < th | dose dependent | sin | 12 | NA |
| 3 | 198.29 | humidity | 50 | 90 | < th | dose independent | sin | 13 | NA |
| 5 | 198.66 | humidity | 50 | 10 | < th | dose independent | rect. | 14 | 2 |
| 10 | 198.95 | humidity | 50 | 90 | > th | dose independent | sin | 13 | NA |
| 12 | 199.42 | humidity | 50 | 10 | < th | dose independent | rect. | 14 | 4 |
| 32 | 199.60 | humidity | 50 | 10 | < th | dose dependent | rect. | 14 | 2 |
| 37 | 199.94 | humidity | 50 | 10 | < th | dose dependent | rect. | 14 | 4 |
| 159 | 200.34 | wind | 1 | 10 | < th | dose independent | rect. | 14 | 2 |
| 161 | 200.34 | wind | 1 | 10 | < th | dose dependent | rect. | 14 | 2 |
| 339 | 204.73 | humidity | 50 | 270 | < th | dose dependent | rect. | 12 | 1 |
| 340 | 204.74 | humidity | 50 | 270 | < th | dose independent | rect. | 12 | 1 |
| 419 | 205.53 | humidity | 50 | 270 | < th | dose dependent | sin | 7 | NA |
| 591 | 208.25 | temperature | 30 | 90 | > th | dose independent | rect. | 15 | 1 |
| 815 | 209.86 | humidity | 50 | 270 | < th | dose independent | no | NA | NA |
| 988 | 209.96 | humidity | 50 | 270 | < th | dose dependent | no | NA | NA |