Os09g0132200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Lipolytic enzyme, G-D-S-L family protein.
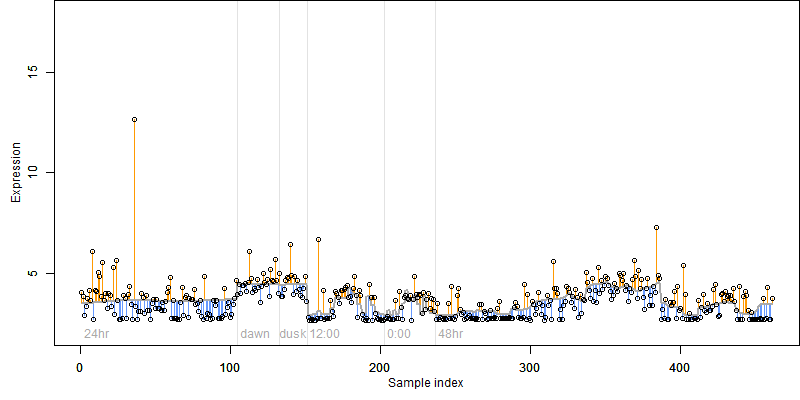

FiT-DB / Search/ Help/ Sample detail
|
Os09g0132200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Lipolytic enzyme, G-D-S-L family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
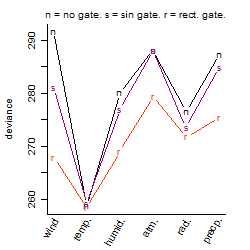
Dependence on each variable

Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 256.74 | 0.753 | 3.62 | 0.401 | 0.117 | 1.6 | -0.892 | 0.739 | -0.224 | 13.1 | 21.58 | 1473 | -- | -- |
| 256.8 | 0.746 | 3.64 | 0.186 | 0.106 | 1.63 | -0.905 | -- | -0.266 | 13.1 | 22.39 | 1489 | -- | -- |
| 259.77 | 0.751 | 3.62 | 0.36 | 0.137 | 1.6 | -- | 0.691 | -0.228 | 12.8 | 22.03 | 1438 | -- | -- |
| 259.96 | 0.751 | 3.64 | 0.153 | 0.145 | 1.63 | -- | -- | -0.268 | 12 | 22.49 | 1444 | -- | -- |
| 260.69 | 0.752 | 3.63 | 0.355 | -- | 1.58 | -- | 0.662 | -0.225 | -- | 22.15 | 1504 | -- | -- |
| 355.3 | 0.884 | 3.56 | 0.779 | 0.005 | -- | -1.1 | -- | 0.105 | 13.9 | -- | -- | -- | -- |
| 359.49 | 0.888 | 3.56 | 0.744 | 0.0908 | -- | -- | -- | 0.0971 | 7.79 | -- | -- | -- | -- |
| 260.9 | 0.752 | 3.64 | 0.151 | -- | 1.61 | -- | -- | -0.26 | -- | 22.52 | 1501 | -- | -- |
| 260.51 | 0.752 | 3.65 | -- | 0.133 | 1.66 | -- | -- | -0.293 | 11.6 | 22.45 | 1464 | -- | -- |
| 359.94 | 0.887 | 3.56 | 0.745 | -- | -- | -- | -- | 0.0984 | -- | -- | -- | -- | -- |
| 374.08 | 0.905 | 3.58 | -- | 0.0973 | -- | -- | -- | 0.0143 | 7.26 | -- | -- | -- | -- |
| 261.42 | 0.753 | 3.65 | -- | -- | 1.65 | -- | -- | -0.293 | -- | 22.46 | 1512 | -- | -- |
| 374.58 | 0.903 | 3.58 | -- | -- | -- | -- | -- | 0.0157 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance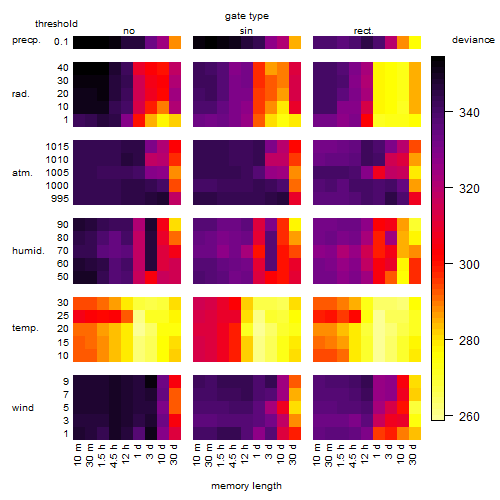
|
Histogram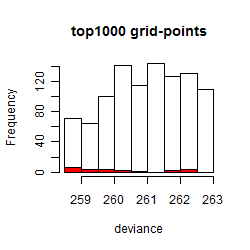 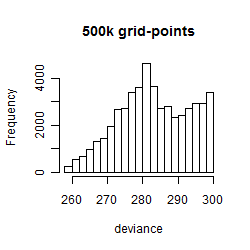
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 258.53 | temperature | 20 | 1440 | > th | dose dependent | rect. | 1 | 21 |
| 7 | 258.60 | temperature | 20 | 1440 | > th | dose dependent | rect. | 12 | 22 |
| 27 | 258.74 | temperature | 20 | 1440 | > th | dose dependent | rect. | 21 | 22 |
| 41 | 258.82 | temperature | 20 | 1440 | > th | dose dependent | rect. | 5 | 23 |
| 42 | 258.83 | temperature | 20 | 1440 | > th | dose dependent | no | NA | NA |
| 44 | 258.84 | temperature | 20 | 1440 | > th | dose dependent | rect. | 8 | 23 |
| 85 | 259.09 | temperature | 20 | 1440 | > th | dose dependent | rect. | 16 | 1 |
| 97 | 259.21 | temperature | 20 | 1440 | > th | dose dependent | sin | 18 | NA |
| 132 | 259.49 | temperature | 20 | 1440 | > th | dose dependent | rect. | 14 | 9 |
| 134 | 259.50 | temperature | 20 | 1440 | > th | dose dependent | rect. | 15 | 7 |
| 201 | 259.88 | temperature | 10 | 1440 | > th | dose dependent | rect. | 16 | 1 |
| 202 | 259.88 | temperature | 25 | 1440 | > th | dose dependent | rect. | 8 | 11 |
| 215 | 259.93 | temperature | 20 | 1440 | > th | dose dependent | rect. | 16 | 3 |
| 244 | 260.05 | temperature | 25 | 1440 | > th | dose dependent | rect. | 10 | 9 |
| 273 | 260.20 | temperature | 20 | 1440 | > th | dose dependent | sin | 4 | NA |
| 444 | 260.77 | temperature | 10 | 1440 | > th | dose dependent | rect. | 16 | 3 |
| 759 | 261.99 | temperature | 20 | 1440 | > th | dose dependent | rect. | 6 | 1 |
| 760 | 262.00 | temperature | 20 | 1440 | > th | dose dependent | rect. | 5 | 4 |
| 864 | 262.42 | temperature | 25 | 1440 | > th | dose dependent | rect. | 12 | 1 |
| 875 | 262.45 | temperature | 30 | 14400 | > th | dose independent | rect. | 15 | 2 |
| 881 | 262.47 | temperature | 30 | 14400 | < th | dose independent | rect. | 15 | 2 |