Os08g0505900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to DNA-damage-repair/toleration protein DRT100 precursor.


FiT-DB / Search/ Help/ Sample detail
|
Os08g0505900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to DNA-damage-repair/toleration protein DRT100 precursor.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

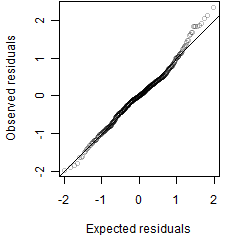
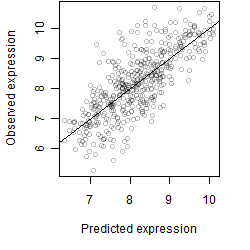
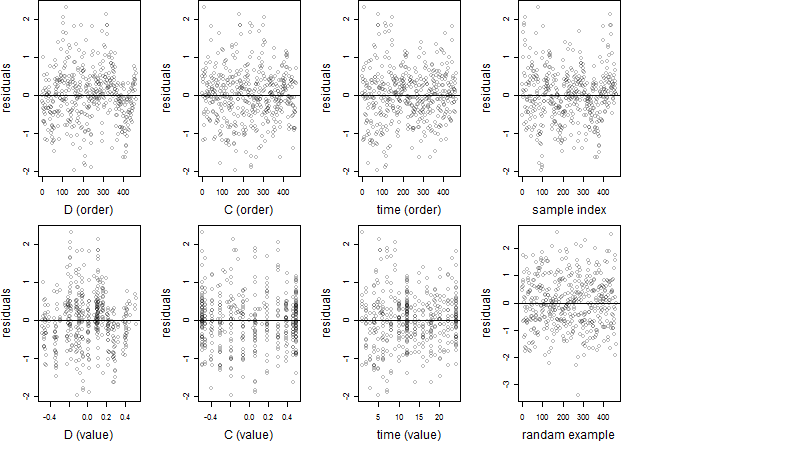
Dependence on each variable

Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 192.21 | 0.651 | 8.37 | -2.56 | 1.55 | 0.558 | 0.015 | -1.71 | -0.955 | 12 | 25.2 | 20 | -- | -- |
| 202.75 | 0.663 | 8.32 | -2.06 | 1.55 | 0.509 | -0.674 | -- | -0.806 | 12 | 25.3 | 19 | -- | -- |
| 191.94 | 0.645 | 8.37 | -2.57 | 1.55 | 0.562 | -- | -1.72 | -0.963 | 12 | 25.3 | 19 | -- | -- |
| 204.67 | 0.671 | 8.32 | -2.05 | 1.56 | 0.48 | -- | -- | -0.787 | 12 | 25.3 | 19 | -- | -- |
| 320.14 | 0.838 | 8.44 | -2.63 | -- | 1.04 | -- | -1.61 | -1.18 | -- | 25.3 | 4 | -- | -- |
| 219.16 | 0.694 | 8.27 | -1.93 | 1.82 | -- | -0.578 | -- | -0.551 | 12.5 | -- | -- | -- | -- |
| 220.59 | 0.696 | 8.27 | -1.96 | 1.82 | -- | -- | -- | -0.554 | 12.5 | -- | -- | -- | -- |
| 330.29 | 0.85 | 8.41 | -2.19 | -- | 1 | -- | -- | -1.06 | -- | 25.4 | 18 | -- | -- |
| 314.1 | 0.83 | 8.25 | -- | 1.67 | 0.321 | -- | -- | -0.485 | 12.3 | 25.3 | 19 | -- | -- |
| 428.94 | 0.968 | 8.3 | -2.02 | -- | -- | -- | -- | -0.585 | -- | -- | -- | -- | -- |
| 321.35 | 0.839 | 8.22 | -- | 1.85 | -- | -- | -- | -0.336 | 12.6 | -- | -- | -- | -- |
| 455.75 | 0.998 | 8.35 | -- | -- | 0.904 | -- | -- | -0.771 | -- | 25.5 | 21 | -- | -- |
| 536.43 | 1.08 | 8.25 | -- | -- | -- | -- | -- | -0.361 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 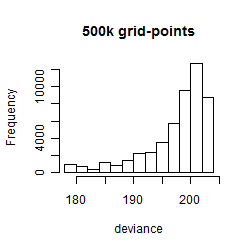
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 178.06 | temperature | 30 | 43200 | > th | dose dependent | rect. | 9 | 3 |
| 35 | 178.10 | temperature | 30 | 43200 | > th | dose dependent | sin | 8 | NA |
| 39 | 178.15 | temperature | 30 | 43200 | < th | dose independent | rect. | 9 | 1 |
| 41 | 178.16 | temperature | 30 | 43200 | < th | dose independent | rect. | 3 | 8 |
| 44 | 178.18 | temperature | 30 | 43200 | < th | dose independent | rect. | 5 | 6 |
| 46 | 178.19 | temperature | 30 | 43200 | > th | dose independent | rect. | 9 | 1 |
| 51 | 178.20 | temperature | 30 | 43200 | < th | dose independent | rect. | 20 | 15 |
| 74 | 178.24 | temperature | 30 | 43200 | < th | dose independent | rect. | 16 | 19 |
| 111 | 178.29 | temperature | 30 | 43200 | > th | dose independent | rect. | 16 | 19 |
| 115 | 178.31 | temperature | 30 | 43200 | > th | dose independent | sin | 9 | NA |
| 116 | 178.31 | temperature | 30 | 43200 | < th | dose independent | sin | 9 | NA |
| 224 | 178.44 | temperature | 30 | 43200 | > th | dose dependent | no | NA | NA |
| 363 | 178.47 | temperature | 25 | 43200 | > th | dose dependent | rect. | 6 | 2 |
| 496 | 178.72 | temperature | 30 | 43200 | < th | dose independent | rect. | 17 | 1 |
| 565 | 178.88 | temperature | 25 | 43200 | > th | dose dependent | rect. | 22 | 1 |
| 572 | 178.94 | temperature | 30 | 43200 | < th | dose independent | rect. | 16 | 6 |
| 573 | 178.94 | temperature | 30 | 43200 | < th | dose independent | rect. | 16 | 15 |
| 577 | 178.95 | temperature | 30 | 43200 | < th | dose independent | rect. | 16 | 3 |
| 587 | 178.97 | temperature | 30 | 43200 | < th | dose independent | rect. | 16 | 12 |
| 594 | 178.99 | temperature | 30 | 43200 | > th | dose independent | rect. | 16 | 3 |
| 756 | 179.56 | temperature | 25 | 43200 | > th | dose independent | rect. | 6 | 1 |
| 763 | 179.60 | temperature | 25 | 43200 | < th | dose independent | rect. | 6 | 1 |
| 840 | 179.99 | temperature | 30 | 43200 | > th | dose independent | no | NA | NA |
| 841 | 179.99 | temperature | 30 | 43200 | > th | dose independent | rect. | 8 | 11 |
| 985 | 180.10 | temperature | 30 | 43200 | < th | dose independent | rect. | 3 | 16 |