Os08g0400000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA).
FiT-DB / Search/ Help/ Sample detail
|
Os08g0400000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA).
|
|
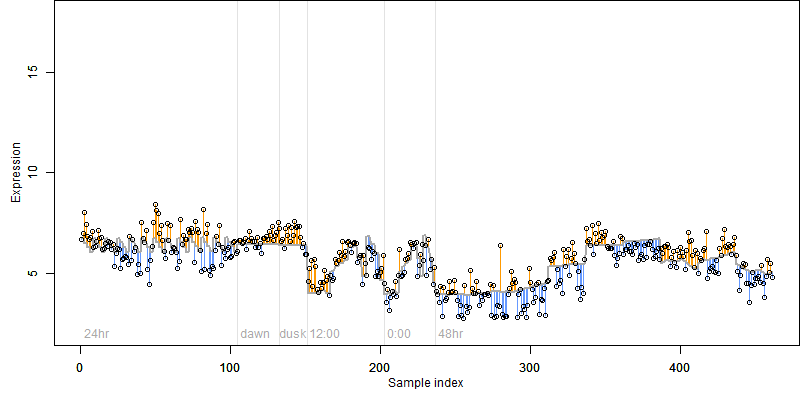
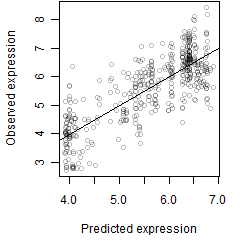
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
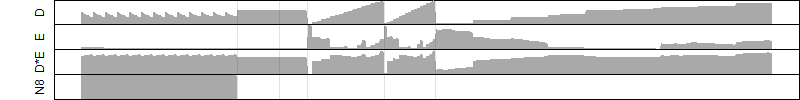
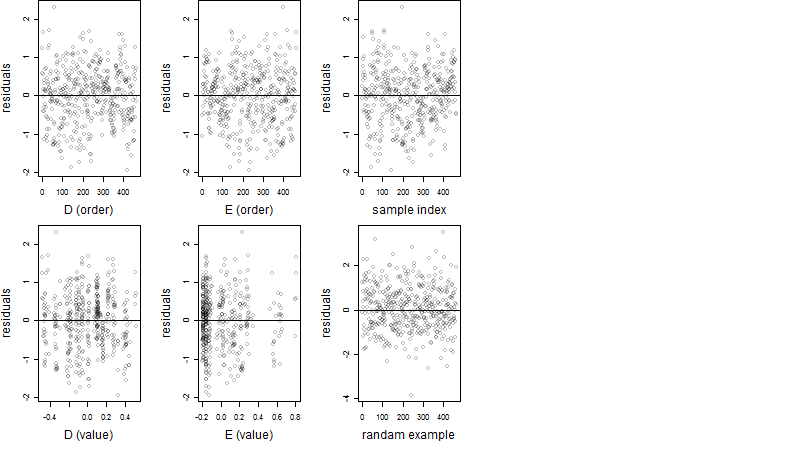
Dependence on each variable

Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 231.26 | 0.714 | 5.2 | 2.57 | 0.0779 | -3.7 | 1.1 | -7.6 | 0.968 | 20.4 | 27.43 | 5834 | -- | -- |
| 265.27 | 0.759 | 5.47 | 1.13 | 0.0414 | -2.86 | -0.997 | -- | 0.628 | 8.67 | 37.18 | 6429 | -- | -- |
| 231.75 | 0.709 | 5.18 | 2.5 | 0.217 | -4.17 | -- | -8.65 | 0.928 | 16.3 | 25.3 | 5746 | -- | -- |
| 266.71 | 0.761 | 5.48 | 1.08 | 0.232 | -2.88 | -- | -- | 0.574 | 15.4 | 186.8 | 6879 | -- | -- |
| 233.99 | 0.712 | 5.18 | 2.46 | -- | -4.2 | -- | -8.71 | 0.912 | -- | 25.3 | 6052 | -- | -- |
| 401.77 | 0.94 | 5.32 | 2.55 | 0.0674 | -- | 1.11 | -- | 1.3 | 21.3 | -- | -- | -- | -- |
| 405.75 | 0.943 | 5.32 | 2.51 | 0.117 | -- | -- | -- | 1.3 | 17.8 | -- | -- | -- | -- |
| 268.92 | 0.764 | 5.47 | 1.1 | -- | -2.86 | -- | -- | 0.624 | -- | 195 | 6402 | -- | -- |
| 241.04 | 0.723 | 5.59 | -- | 0.163 | -3.05 | -- | -- | 0.0887 | 15.6 | 163.2 | 32226 | -- | -- |
| 406.46 | 0.942 | 5.32 | 2.51 | -- | -- | -- | -- | 1.3 | -- | -- | -- | -- | -- |
| 571.86 | 1.12 | 5.38 | -- | 0.0844 | -- | -- | -- | 1.02 | 19.3 | -- | -- | -- | -- |
| 242.49 | 0.725 | 5.59 | -- | -- | -3.03 | -- | -- | 0.0872 | -- | 169.1 | 32153 | -- | -- |
| 572.23 | 1.12 | 5.38 | -- | -- | -- | -- | -- | 1.02 | -- | -- | -- | -- | -- |
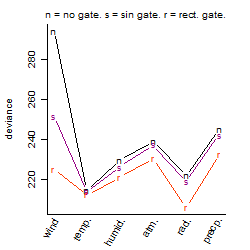
Results of the grid search
Summarized heatmap of deviance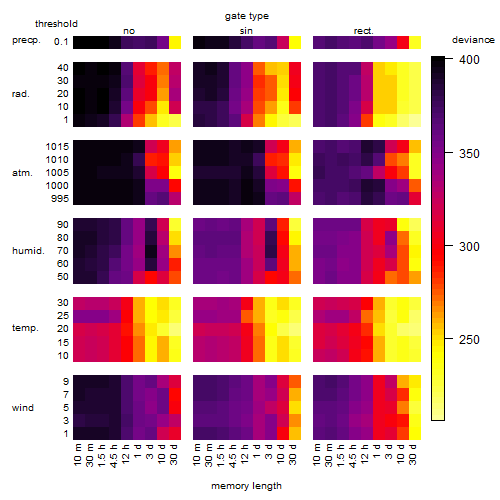
|
Histogram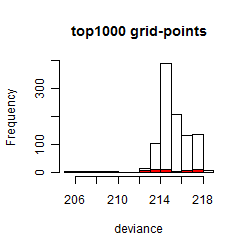 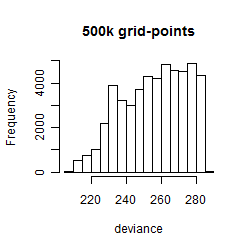
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 205.84 | radiation | 1 | 43200 | < th | dose dependent | rect. | 4 | 1 |
| 6 | 208.38 | radiation | 1 | 43200 | < th | dose dependent | rect. | 21 | 8 |
| 11 | 212.02 | temperature | 20 | 43200 | > th | dose independent | rect. | 6 | 1 |
| 12 | 212.04 | temperature | 20 | 43200 | < th | dose independent | rect. | 5 | 2 |
| 13 | 212.17 | temperature | 20 | 43200 | < th | dose independent | rect. | 1 | 1 |
| 16 | 212.33 | temperature | 20 | 43200 | < th | dose dependent | rect. | 4 | 1 |
| 18 | 212.58 | temperature | 20 | 43200 | < th | dose independent | rect. | 23 | 1 |
| 20 | 212.80 | temperature | 20 | 43200 | < th | dose independent | rect. | 23 | 3 |
| 27 | 213.26 | temperature | 20 | 43200 | > th | dose independent | rect. | 23 | 1 |
| 32 | 213.42 | temperature | 20 | 43200 | < th | dose independent | rect. | 21 | 15 |
| 36 | 213.49 | temperature | 20 | 43200 | < th | dose independent | rect. | 21 | 19 |
| 40 | 213.52 | temperature | 20 | 43200 | < th | dose independent | rect. | 21 | 10 |
| 72 | 213.70 | temperature | 20 | 43200 | < th | dose independent | sin | 8 | NA |
| 98 | 213.88 | temperature | 20 | 43200 | > th | dose independent | rect. | 1 | 1 |
| 120 | 213.99 | temperature | 20 | 43200 | > th | dose independent | rect. | 23 | 3 |
| 124 | 214.00 | temperature | 20 | 43200 | < th | dose independent | rect. | 14 | 17 |
| 144 | 214.09 | temperature | 20 | 43200 | > th | dose independent | rect. | 21 | 15 |
| 151 | 214.11 | temperature | 20 | 43200 | > th | dose independent | rect. | 20 | 11 |
| 170 | 214.17 | temperature | 20 | 43200 | > th | dose independent | rect. | 21 | 18 |
| 221 | 214.27 | temperature | 20 | 43200 | < th | dose independent | rect. | 5 | 22 |
| 222 | 214.28 | temperature | 20 | 43200 | > th | dose independent | sin | 9 | NA |
| 314 | 214.48 | temperature | 20 | 43200 | > th | dose independent | rect. | 14 | 17 |
| 318 | 214.49 | temperature | 20 | 43200 | < th | dose independent | no | NA | NA |
| 354 | 214.60 | temperature | 20 | 43200 | > th | dose independent | rect. | 1 | 7 |
| 390 | 214.69 | temperature | 20 | 43200 | < th | dose independent | rect. | 1 | 23 |
| 403 | 214.73 | temperature | 20 | 43200 | > th | dose independent | rect. | 5 | 22 |
| 501 | 214.94 | temperature | 20 | 43200 | > th | dose independent | no | NA | NA |
| 527 | 215.04 | temperature | 20 | 43200 | > th | dose independent | rect. | 1 | 23 |
| 771 | 216.31 | radiation | 1 | 14400 | < th | dose independent | rect. | 18 | 11 |
| 790 | 216.48 | radiation | 1 | 43200 | > th | dose independent | rect. | 18 | 1 |
| 803 | 216.50 | temperature | 25 | 43200 | > th | dose independent | rect. | 17 | 1 |
| 816 | 216.61 | temperature | 25 | 43200 | < th | dose independent | rect. | 17 | 1 |
| 844 | 216.83 | radiation | 1 | 43200 | < th | dose dependent | rect. | 6 | 23 |
| 886 | 217.20 | radiation | 1 | 14400 | < th | dose dependent | rect. | 18 | 1 |
| 899 | 217.29 | radiation | 1 | 43200 | < th | dose independent | rect. | 18 | 7 |
| 905 | 217.31 | radiation | 1 | 43200 | < th | dose independent | rect. | 18 | 1 |
| 908 | 217.32 | temperature | 20 | 43200 | < th | dose dependent | sin | 8 | NA |
| 912 | 217.33 | radiation | 1 | 43200 | < th | dose dependent | rect. | 6 | 13 |
| 916 | 217.35 | radiation | 1 | 14400 | < th | dose dependent | rect. | 18 | 5 |
| 919 | 217.35 | radiation | 1 | 43200 | < th | dose dependent | rect. | 6 | 17 |
| 920 | 217.35 | radiation | 1 | 43200 | < th | dose dependent | rect. | 8 | 21 |
| 947 | 217.57 | radiation | 1 | 43200 | < th | dose dependent | rect. | 6 | 21 |
| 978 | 217.85 | radiation | 1 | 43200 | < th | dose dependent | rect. | 8 | 11 |
| 982 | 217.88 | radiation | 1 | 43200 | < th | dose dependent | rect. | 8 | 15 |