Os08g0395800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF247, plant family protein.
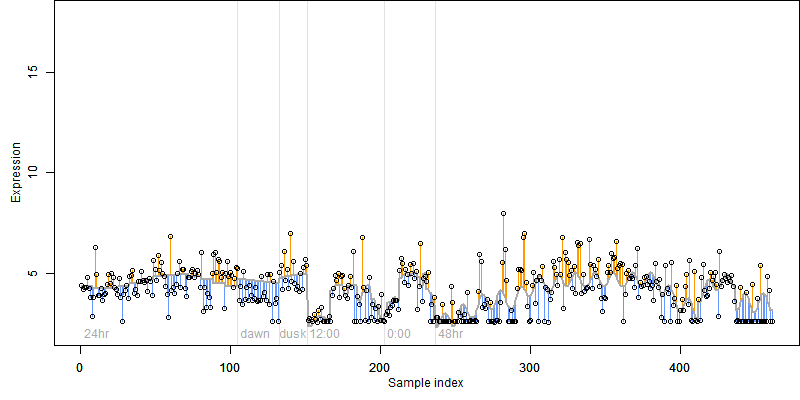

FiT-DB / Search/ Help/ Sample detail
|
Os08g0395800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF247, plant family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
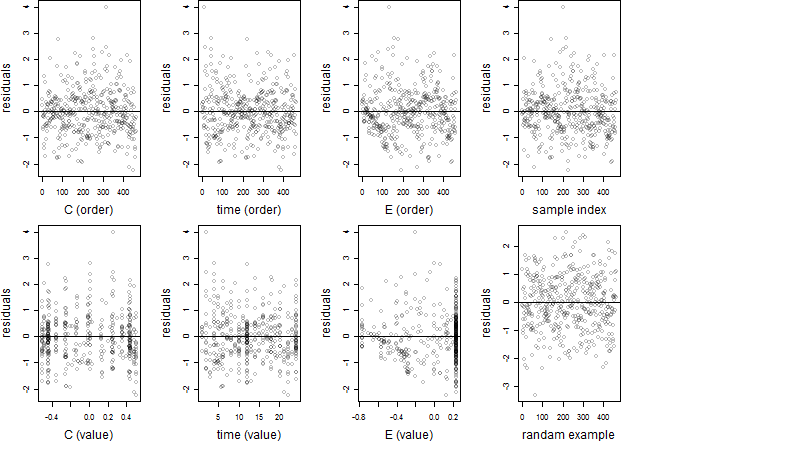
Dependence on each variable

Residual plot
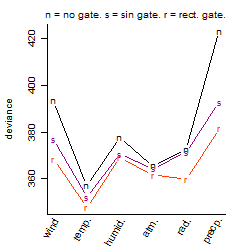
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 349.01 | 0.878 | 4.21 | -0.728 | 0.698 | 2.22 | -0.217 | -0.3 | -0.205 | 22.5 | 21.8 | 3935 | -- | -- |
| 349.11 | 0.87 | 4.2 | -0.633 | 0.69 | 2.24 | -0.237 | -- | -0.183 | 22.6 | 21.76 | 3845 | -- | -- |
| 349.18 | 0.87 | 4.21 | -0.721 | 0.697 | 2.23 | -- | -0.304 | -0.205 | 22.4 | 21.78 | 3895 | -- | -- |
| 349.33 | 0.87 | 4.2 | -0.623 | 0.684 | 2.24 | -- | -- | -0.181 | 22.5 | 21.7 | 3828 | -- | -- |
| 377.65 | 0.905 | 4.2 | -0.722 | -- | 2.29 | -- | -0.25 | -0.218 | -- | 21.7 | 3793 | -- | -- |
| 485.55 | 1.03 | 4.02 | 0.668 | 0.802 | -- | -0.252 | -- | 0.61 | 22.9 | -- | -- | -- | -- |
| 485.8 | 1.03 | 4.02 | 0.679 | 0.804 | -- | -- | -- | 0.611 | 22.8 | -- | -- | -- | -- |
| 372.17 | 0.899 | 4.17 | -0.42 | -- | 2.25 | -- | -- | -0.122 | -- | 21.76 | 2502 | -- | -- |
| 346.44 | 0.867 | 4.2 | -- | 0.705 | 2.02 | -- | -- | -0.101 | 22.1 | 22 | 2590 | -- | -- |
| 525.83 | 1.07 | 4 | 0.696 | -- | -- | -- | -- | 0.616 | -- | -- | -- | -- | -- |
| 497.96 | 1.04 | 4.04 | -- | 0.809 | -- | -- | -- | 0.535 | 22.9 | -- | -- | -- | -- |
| 371.77 | 0.898 | 4.16 | -- | -- | 2.05 | -- | -- | -0.0805 | -- | 22.18 | 2467 | -- | -- |
| 538.6 | 1.08 | 4.02 | -- | -- | -- | -- | -- | 0.539 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance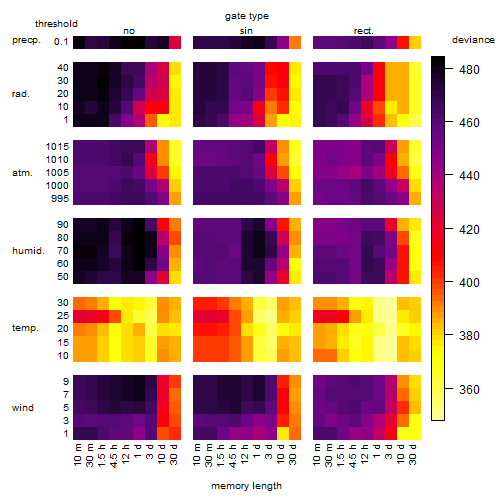
|
Histogram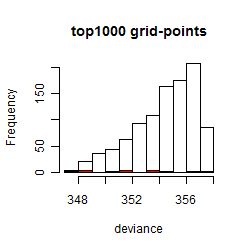 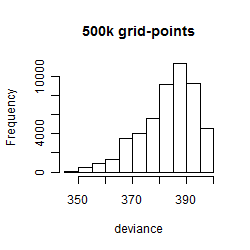
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 347.76 | temperature | 25 | 4320 | < th | dose dependent | rect. | 21 | 5 |
| 14 | 348.62 | temperature | 15 | 4320 | > th | dose dependent | rect. | 23 | 3 |
| 16 | 348.77 | temperature | 25 | 4320 | < th | dose dependent | rect. | 1 | 1 |
| 19 | 348.84 | temperature | 15 | 4320 | > th | dose dependent | rect. | 1 | 1 |
| 85 | 350.54 | temperature | 15 | 4320 | > th | dose dependent | rect. | 3 | 1 |
| 102 | 350.97 | temperature | 20 | 4320 | > th | dose independent | rect. | 22 | 1 |
| 107 | 351.04 | temperature | 30 | 1440 | < th | dose dependent | rect. | 3 | 1 |
| 117 | 351.34 | temperature | 20 | 4320 | < th | dose independent | rect. | 22 | 1 |
| 166 | 351.97 | temperature | 25 | 4320 | < th | dose dependent | sin | 13 | NA |
| 297 | 353.29 | temperature | 20 | 4320 | > th | dose independent | rect. | 17 | 11 |
| 321 | 353.47 | temperature | 20 | 4320 | < th | dose independent | rect. | 18 | 9 |
| 345 | 353.76 | temperature | 20 | 4320 | > th | dose independent | rect. | 19 | 7 |
| 366 | 353.96 | temperature | 20 | 4320 | < th | dose independent | rect. | 20 | 6 |
| 417 | 354.25 | temperature | 20 | 4320 | > th | dose independent | sin | 14 | NA |
| 453 | 354.47 | temperature | 20 | 4320 | < th | dose independent | sin | 13 | NA |
| 730 | 356.11 | temperature | 15 | 4320 | > th | dose dependent | sin | 11 | NA |
| 939 | 357.15 | temperature | 25 | 4320 | < th | dose dependent | no | NA | NA |