Os08g0249600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Diacylglycerol kinase-like protein.

FiT-DB / Search/ Help/ Sample detail
|
Os08g0249600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Diacylglycerol kinase-like protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

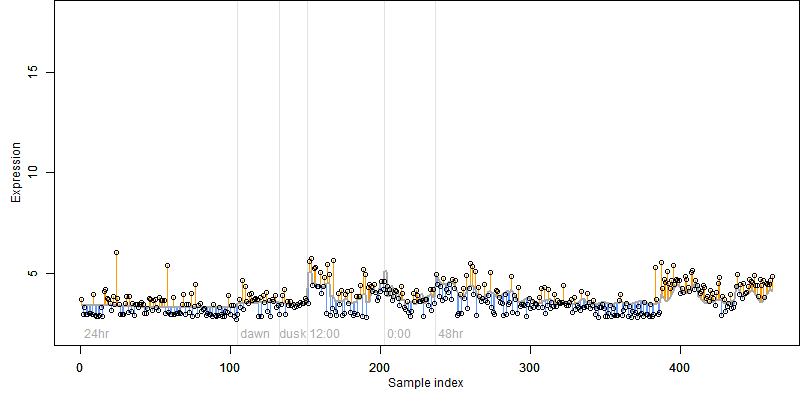
 __
__

Dependence on each variable

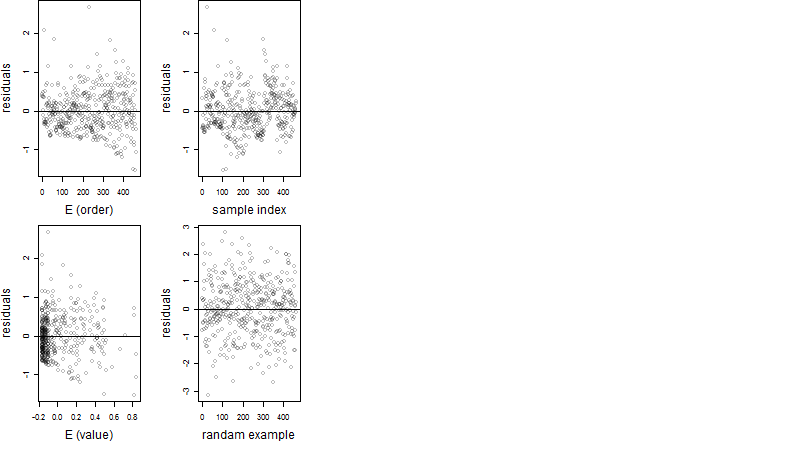
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 126.11 | 0.528 | 3.7 | 0.416 | 0.158 | 1.85 | -0.0292 | 0.315 | -0.107 | 17.1 | 25.19 | 650 | -- | -- |
| 125.97 | 0.523 | 3.7 | 0.463 | 0.227 | 1.83 | -0.045 | -- | -0.0853 | 17.1 | 25.85 | 583 | -- | -- |
| 125.94 | 0.523 | 3.7 | 0.434 | 0.228 | 1.84 | -- | 0.177 | -0.0875 | 17.1 | 25.77 | 585 | -- | -- |
| 125.97 | 0.523 | 3.7 | 0.468 | 0.232 | 1.82 | -- | -- | -0.0789 | 17 | 25.84 | 581 | -- | -- |
| 126.89 | 0.525 | 3.71 | 0.407 | -- | 1.84 | -- | 0.421 | -0.116 | -- | 25.19 | 831 | -- | -- |
| 175.5 | 0.621 | 3.78 | -0.0254 | 0.24 | -- | -0.162 | -- | -0.479 | 11.7 | -- | -- | -- | -- |
| 175.62 | 0.621 | 3.78 | -0.032 | 0.241 | -- | -- | -- | -0.48 | 11.7 | -- | -- | -- | -- |
| 127.07 | 0.525 | 3.69 | 0.486 | -- | 1.8 | -- | -- | -0.088 | -- | 25.7 | 839 | -- | -- |
| 131.01 | 0.533 | 3.71 | -- | 0.216 | 1.63 | -- | -- | -0.17 | 16.3 | 25.7 | 552 | -- | -- |
| 179.26 | 0.626 | 3.78 | -0.0389 | -- | -- | -- | -- | -0.483 | -- | -- | -- | -- | -- |
| 175.64 | 0.62 | 3.78 | -- | 0.241 | -- | -- | -- | -0.476 | 11.7 | -- | -- | -- | -- |
| 132.3 | 0.536 | 3.72 | -- | -- | 1.6 | -- | -- | -0.188 | -- | 25.23 | 724 | -- | -- |
| 179.3 | 0.625 | 3.78 | -- | -- | -- | -- | -- | -0.479 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 122.63 | temperature | 30 | 720 | < th | dose dependent | rect. | 16 | 19 |
| 2 | 122.68 | temperature | 30 | 720 | < th | dose dependent | rect. | 17 | 17 |
| 5 | 122.71 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 18 |
| 9 | 122.79 | temperature | 10 | 720 | > th | dose dependent | rect. | 18 | 15 |
| 18 | 123.65 | temperature | 15 | 720 | > th | dose dependent | sin | 10 | NA |
| 32 | 123.88 | temperature | 30 | 720 | < th | dose dependent | sin | 10 | NA |
| 131 | 125.64 | temperature | 15 | 1440 | > th | dose dependent | rect. | 23 | 10 |
| 144 | 125.69 | temperature | 25 | 1440 | < th | dose dependent | rect. | 4 | 1 |
| 148 | 125.70 | temperature | 30 | 1440 | < th | dose dependent | rect. | 20 | 11 |
| 165 | 125.75 | temperature | 30 | 1440 | < th | dose dependent | rect. | 23 | 10 |
| 231 | 125.95 | temperature | 15 | 1440 | > th | dose dependent | rect. | 4 | 1 |
| 283 | 126.09 | temperature | 30 | 1440 | < th | dose dependent | rect. | 4 | 3 |
| 296 | 126.12 | temperature | 15 | 1440 | > th | dose dependent | rect. | 4 | 3 |
| 365 | 126.31 | temperature | 25 | 720 | < th | dose dependent | rect. | 19 | 23 |
| 373 | 126.34 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 418 | 126.43 | temperature | 25 | 720 | < th | dose dependent | rect. | 6 | 23 |
| 437 | 126.47 | temperature | 25 | 720 | < th | dose dependent | rect. | 3 | 23 |
| 565 | 126.79 | temperature | 25 | 720 | < th | dose dependent | rect. | 24 | 23 |
| 599 | 126.89 | temperature | 10 | 720 | > th | dose dependent | rect. | 11 | 22 |
| 690 | 127.09 | temperature | 30 | 1440 | < th | dose dependent | rect. | 1 | 1 |