Os08g0248800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Aspartate carbamoyltransferase 3, chloroplast precursor (EC 2.1.3.2) (Aspartate transcarbamylase 3) (ATCase 3).


FiT-DB / Search/ Help/ Sample detail
|
Os08g0248800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Aspartate carbamoyltransferase 3, chloroplast precursor (EC 2.1.3.2) (Aspartate transcarbamylase 3) (ATCase 3).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

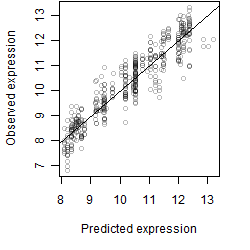
Dependence on each variable

Residual plot
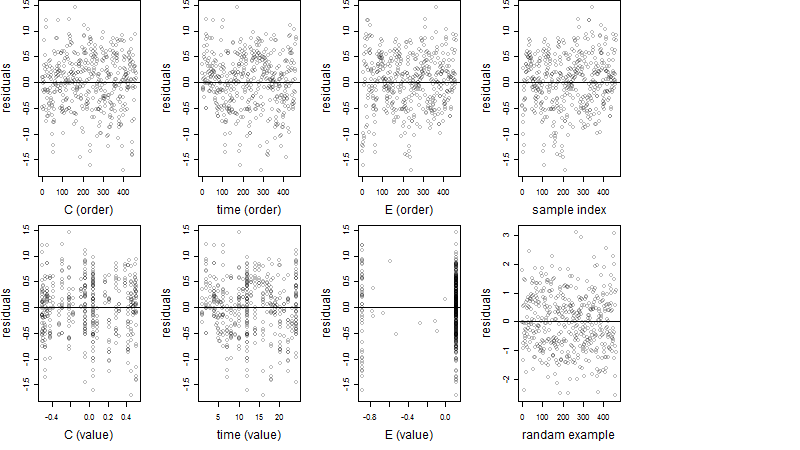
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 121.91 | 0.514 | 10.5 | 0.171 | 4.06 | -0.919 | 1.2 | -0.457 | -0.242 | 17.7 | 20 | 30 | -- | -- |
| 122.9 | 0.516 | 10.4 | 0.244 | 4.05 | -0.891 | 1.04 | -- | -0.226 | 17.7 | 20.02 | 36 | -- | -- |
| 125.06 | 0.521 | 10.4 | 0.191 | 4.06 | -0.905 | -- | -0.281 | -0.233 | 17.7 | 20.1 | 49 | -- | -- |
| 125.43 | 0.522 | 10.4 | 0.243 | 4.06 | -0.886 | -- | -- | -0.223 | 17.7 | 20.01 | 49 | -- | -- |
| 963.59 | 1.45 | 10.4 | 0.0434 | -- | -0.552 | -- | -0.0259 | -0.398 | -- | 19.3 | 18 | -- | -- |
| 153.47 | 0.581 | 10.5 | 0.0135 | 3.97 | -- | 0.804 | -- | -0.385 | 17.9 | -- | -- | -- | -- |
| 155.05 | 0.583 | 10.5 | 0.00768 | 3.97 | -- | -- | -- | -0.381 | 17.9 | -- | -- | -- | -- |
| 963.6 | 1.45 | 10.4 | 0.046 | -- | -0.548 | -- | -- | -0.398 | -- | 19.12 | 31 | -- | -- |
| 126.92 | 0.525 | 10.4 | -- | 4.06 | -0.843 | -- | -- | -0.245 | 17.7 | 20 | 49 | -- | -- |
| 973.14 | 1.46 | 10.4 | -0.113 | -- | -- | -- | -- | -0.474 | -- | -- | -- | -- | -- |
| 155.05 | 0.582 | 10.5 | -- | 3.97 | -- | -- | -- | -0.382 | 17.9 | -- | -- | -- | -- |
| 963.65 | 1.45 | 10.4 | -- | -- | -0.538 | -- | -- | -0.404 | -- | 19.12 | 31 | -- | -- |
| 973.48 | 1.46 | 10.4 | -- | -- | -- | -- | -- | -0.461 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram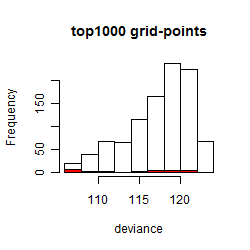 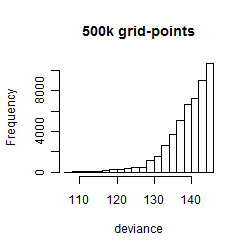
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 106.96 | temperature | 20 | 10 | < th | dose independent | rect. | 23 | 11 |
| 9 | 107.18 | temperature | 20 | 270 | > th | dose independent | rect. | 17 | 14 |
| 10 | 107.53 | temperature | 20 | 30 | > th | dose independent | sin | 9 | NA |
| 12 | 107.88 | temperature | 20 | 10 | < th | dose independent | rect. | 23 | 9 |
| 15 | 107.96 | temperature | 20 | 90 | > th | dose independent | rect. | 19 | 14 |
| 29 | 108.44 | temperature | 25 | 10 | < th | dose dependent | rect. | 23 | 8 |
| 40 | 108.78 | temperature | 30 | 270 | < th | dose dependent | rect. | 21 | 7 |
| 58 | 110.07 | temperature | 20 | 30 | < th | dose independent | sin | 9 | NA |
| 215 | 114.86 | temperature | 25 | 10 | < th | dose dependent | sin | 9 | NA |
| 264 | 115.53 | temperature | 15 | 270 | > th | dose dependent | rect. | 18 | 13 |
| 364 | 116.89 | temperature | 20 | 720 | > th | dose dependent | rect. | 20 | 4 |
| 380 | 117.07 | temperature | 20 | 30 | < th | dose independent | rect. | 14 | 20 |
| 461 | 117.90 | temperature | 25 | 720 | < th | dose dependent | rect. | 21 | 2 |
| 499 | 118.18 | temperature | 20 | 30 | > th | dose independent | rect. | 15 | 23 |
| 584 | 119.04 | temperature | 20 | 10 | > th | dose dependent | rect. | 21 | 12 |
| 587 | 119.08 | temperature | 15 | 10 | > th | dose dependent | sin | 9 | NA |
| 816 | 121.01 | temperature | 20 | 720 | > th | dose independent | rect. | 19 | 5 |
| 858 | 121.48 | temperature | 20 | 30 | > th | dose independent | rect. | 17 | 23 |
| 861 | 121.50 | temperature | 20 | 30 | > th | dose independent | rect. | 13 | 23 |
| 914 | 121.91 | temperature | 20 | 30 | > th | dose independent | no | NA | NA |