Os08g0155700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to RNA polymerase II largest subunit (Fragment).


FiT-DB / Search/ Help/ Sample detail
|
Os08g0155700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to RNA polymerase II largest subunit (Fragment).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
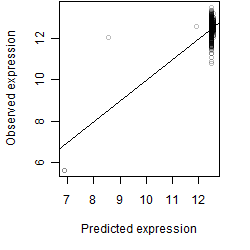
Dependence on each variable

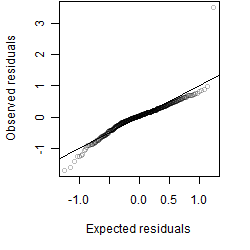
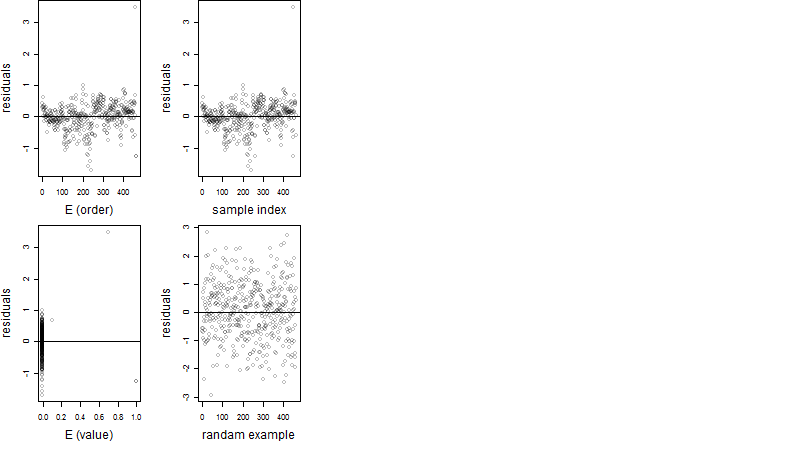
Residual plot
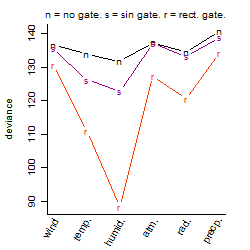
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = < th, dose dependency = dose independent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 88.17 | 0.441 | 11.5 | 9.96 | 0.393 | -64.7 | -0.171 | 623 | 0.145 | 3.46 | 59.99 | 31 | 17 | 0.999 |
| 115.76 | 0.505 | 12.4 | 0.546 | 0.401 | -2 | -0.18 | -- | 0.148 | 3.59 | 59.99 | 31 | 17 | 0.999 |
| 88.24 | 0.437 | 11.5 | 9.97 | 0.392 | -64.7 | -- | 623 | 0.146 | 3.37 | 60.02 | 32 | 17 | 0.999 |
| 104.65 | 0.476 | 12.4 | 0.541 | 0.229 | -3 | -- | -- | 0.117 | 1.95 | 53 | 45 | 17.3 | 0.683 |
| 96.67 | 0.458 | 11.5 | 9.98 | -- | -64.9 | -- | 623 | 0.153 | -- | 60.04 | 32 | 17 | 0.991 |
| 142 | 0.559 | 12.4 | 0.505 | 0.52 | -- | -0.0142 | -- | 0.181 | 4.1 | -- | -- | -- | -- |
| 142 | 0.558 | 12.4 | 0.505 | 0.52 | -- | -- | -- | 0.181 | 4.1 | -- | -- | -- | -- |
| 92.51 | 0.448 | 12.4 | 1.06 | -- | -4.75 | -- | -- | 0.246 | -- | 54 | 21 | 17.7 | 5.21 |
| 106.81 | 0.481 | 12.5 | -- | 0.372 | -3.33 | -- | -- | 0.0861 | 3.3 | 54.18 | 40 | 17.3 | 0.567 |
| 156.69 | 0.585 | 12.4 | 0.526 | -- | -- | -- | -- | 0.195 | -- | -- | -- | -- | -- |
| 148.72 | 0.57 | 12.4 | -- | 0.53 | -- | -- | -- | 0.124 | 4.08 | -- | -- | -- | -- |
| 86.43 | 0.433 | 12.5 | -- | -- | -5.59 | -- | -- | 0.0924 | -- | 54.58 | 17 | 17.7 | 4.61 |
| 163.99 | 0.598 | 12.4 | -- | -- | -- | -- | -- | 0.136 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 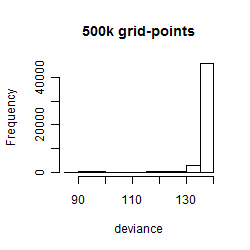
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 88.21 | humidity | 60 | 30 | < th | dose independent | rect. | 17 | 1 |
| 3 | 91.92 | humidity | 60 | 30 | < th | dose dependent | rect. | 17 | 1 |
| 253 | 110.90 | temperature | 30 | 30 | > th | dose independent | rect. | 17 | 1 |
| 255 | 113.48 | temperature | 30 | 30 | > th | dose dependent | rect. | 17 | 1 |
| 379 | 120.53 | radiation | 10 | 90 | > th | dose independent | rect. | 17 | 1 |
| 394 | 120.95 | radiation | 10 | 90 | > th | dose dependent | rect. | 17 | 1 |
| 449 | 122.86 | humidity | 60 | 10 | < th | dose independent | sin | 11 | NA |
| 450 | 122.93 | temperature | 30 | 10 | > th | dose independent | rect. | 14 | 4 |
| 542 | 125.04 | humidity | 50 | 90 | > th | dose independent | sin | 9 | NA |
| 570 | 125.73 | humidity | 60 | 10 | > th | dose independent | sin | 11 | NA |
| 607 | 126.19 | temperature | 30 | 30 | > th | dose dependent | sin | 11 | NA |
| 610 | 126.23 | humidity | 50 | 90 | < th | dose independent | sin | 9 | NA |
| 646 | 126.84 | humidity | 50 | 90 | < th | dose dependent | sin | 9 | NA |
| 679 | 127.39 | humidity | 60 | 90 | < th | dose dependent | sin | 11 | NA |
| 680 | 127.40 | atmosphere | 1005 | 30 | < th | dose independent | rect. | 17 | 1 |
| 764 | 128.40 | temperature | 30 | 270 | > th | dose dependent | rect. | 14 | 1 |
| 811 | 128.85 | temperature | 30 | 10 | > th | dose independent | sin | 12 | NA |