Os08g0153800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF599 family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os08g0153800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF599 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
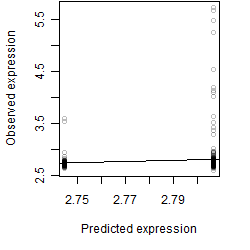
Dependence on each variable

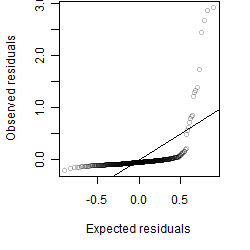
Residual plot

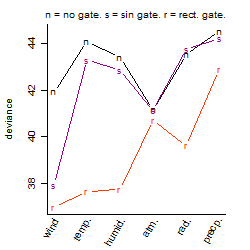
Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 41.19 | 0.299 | 2.8 | 0.0319 | 0.0286 | 0.606 | 0.333 | 2.01 | -0.0523 | 7.33 | 1015 | 10 | -- | -- |
| 43.29 | 0.309 | 2.8 | 0.0631 | 0.00677 | 0.642 | 0.343 | -- | -0.0465 | 5.44 | 1015 | 10 | -- | -- |
| 41.44 | 0.302 | 2.8 | 0.0411 | 0.0394 | 0.619 | -- | 1.95 | -0.052 | 10.4 | 1014 | 206 | -- | -- |
| 43.48 | 0.307 | 2.8 | 0.0484 | 0.0394 | 0.699 | -- | -- | -0.0497 | 10.2 | 1015 | 250 | -- | -- |
| 41.51 | 0.3 | 2.81 | 0.0456 | -- | 0.591 | -- | 1.96 | -0.0526 | -- | 1014 | 278 | -- | -- |
| 44.92 | 0.314 | 2.8 | 0.0631 | 0.00915 | -- | 0.341 | -- | -0.0538 | 5.33 | -- | -- | -- | -- |
| 45.19 | 0.315 | 2.8 | 0.0641 | 0.0207 | -- | -- | -- | -0.0552 | 8.99 | -- | -- | -- | -- |
| 43.57 | 0.307 | 2.8 | 0.0642 | -- | 0.648 | -- | -- | -0.0478 | -- | 1015 | 275 | -- | -- |
| 43.58 | 0.307 | 2.8 | -- | 0.0394 | 0.667 | -- | -- | -0.0547 | 10.4 | 1015 | 260 | -- | -- |
| 45.22 | 0.314 | 2.81 | 0.0641 | -- | -- | -- | -- | -0.0551 | -- | -- | -- | -- | -- |
| 45.3 | 0.315 | 2.81 | -- | 0.0208 | -- | -- | -- | -0.0623 | 8.75 | -- | -- | -- | -- |
| 43.68 | 0.308 | 2.81 | -- | -- | 0.648 | -- | -- | -0.055 | -- | 1015 | 258 | -- | -- |
| 45.32 | 0.314 | 2.81 | -- | -- | -- | -- | -- | -0.0622 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 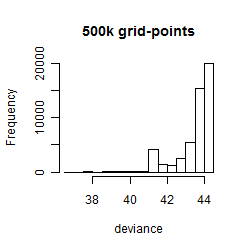
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 36.99 | wind | 9 | 30 | > th | dose dependent | rect. | 11 | 1 |
| 36 | 37.66 | temperature | 20 | 10 | < th | dose independent | rect. | 19 | 1 |
| 45 | 37.66 | temperature | 20 | 10 | < th | dose dependent | rect. | 19 | 1 |
| 64 | 37.75 | humidity | 90 | 90 | > th | dose dependent | rect. | 18 | 2 |
| 66 | 37.77 | humidity | 90 | 90 | > th | dose independent | rect. | 18 | 1 |
| 69 | 37.87 | wind | 9 | 30 | > th | dose independent | rect. | 11 | 1 |
| 71 | 37.93 | wind | 9 | 30 | > th | dose dependent | sin | 6 | NA |
| 127 | 38.65 | wind | 9 | 30 | < th | dose independent | sin | 6 | NA |
| 186 | 39.02 | wind | 9 | 30 | > th | dose independent | sin | 6 | NA |
| 255 | 39.55 | wind | 3 | 10 | < th | dose dependent | rect. | 20 | 1 |
| 258 | 39.62 | radiation | 10 | 10 | < th | dose independent | rect. | 12 | 1 |
| 320 | 40.22 | humidity | 90 | 270 | > th | dose dependent | rect. | 16 | 4 |
| 368 | 40.49 | wind | 3 | 10 | < th | dose independent | rect. | 19 | 2 |
| 401 | 40.65 | humidity | 90 | 270 | > th | dose dependent | rect. | 12 | 8 |
| 410 | 40.71 | atmosphere | 1015 | 720 | < th | dose independent | rect. | 16 | 9 |
| 443 | 40.85 | atmosphere | 1015 | 720 | < th | dose independent | rect. | 16 | 17 |
| 445 | 40.86 | atmosphere | 1015 | 720 | < th | dose independent | rect. | 13 | 15 |
| 497 | 40.99 | temperature | 20 | 90 | < th | dose dependent | rect. | 12 | 8 |
| 499 | 41.00 | atmosphere | 1015 | 720 | < th | dose independent | rect. | 5 | 23 |
| 513 | 41.02 | atmosphere | 1015 | 720 | < th | dose independent | rect. | 21 | 7 |
| 538 | 41.13 | atmosphere | 1015 | 10 | < th | dose independent | sin | 7 | NA |
| 543 | 41.13 | atmosphere | 1015 | 10 | < th | dose independent | sin | 18 | NA |
| 575 | 41.15 | atmosphere | 1015 | 1440 | > th | dose independent | rect. | 22 | 2 |
| 586 | 41.16 | atmosphere | 1015 | 1440 | < th | dose independent | rect. | 22 | 2 |
| 636 | 41.19 | atmosphere | 1015 | 10 | > th | dose independent | sin | 7 | NA |
| 732 | 41.19 | atmosphere | 1015 | 10 | > th | dose independent | no | NA | NA |
| 739 | 41.19 | atmosphere | 1015 | 10 | < th | dose independent | no | NA | NA |
| 743 | 41.19 | atmosphere | 1015 | 270 | > th | dose independent | rect. | 20 | 1 |
| 749 | 41.19 | atmosphere | 1015 | 10 | > th | dose independent | rect. | 23 | 1 |
| 855 | 41.19 | atmosphere | 1015 | 1440 | < th | dose independent | rect. | 22 | 5 |