Os07g0680300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Ubiquinol-cytochrome c reductase complex 6.7 kDa protein (EC 1.10.2.2) (CR6).


FiT-DB / Search/ Help/ Sample detail
|
Os07g0680300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Ubiquinol-cytochrome c reductase complex 6.7 kDa protein (EC 1.10.2.2) (CR6).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22.08 | 0.221 | 11.6 | -0.56 | 0.259 | 0.786 | -0.0177 | 0.26 | -0.0386 | 3.41 | 14.76 | 3890 | -- | -- |
| 22.11 | 0.219 | 11.6 | -0.637 | 0.26 | 0.763 | 0.00069 | -- | -0.0581 | 3.26 | 15.62 | 3975 | -- | -- |
| 22.07 | 0.219 | 11.6 | -0.551 | 0.257 | 0.762 | -- | 0.265 | -0.0362 | 3.42 | 15.79 | 3897 | -- | -- |
| 22.08 | 0.219 | 11.6 | -0.594 | 0.269 | 0.711 | -- | -- | -0.0447 | 3.32 | 16.88 | 3897 | -- | -- |
| 25.51 | 0.235 | 11.6 | -0.579 | -- | 0.758 | -- | -0.00648 | -0.0685 | -- | 16.22 | 2584 | -- | -- |
| 32.75 | 0.268 | 11.6 | -0.278 | 0.311 | -- | 0.0718 | -- | 0.0963 | 2.82 | -- | -- | -- | -- |
| 32.77 | 0.268 | 11.6 | -0.28 | 0.312 | -- | -- | -- | 0.0958 | 2.89 | -- | -- | -- | -- |
| 25.49 | 0.235 | 11.6 | -0.562 | -- | 0.732 | -- | -- | -0.0552 | -- | 17 | 2585 | -- | -- |
| 28.57 | 0.249 | 11.6 | -- | 0.31 | 0.468 | -- | -- | 0.0462 | 4.1 | 21.35 | 2210 | -- | -- |
| 38.38 | 0.289 | 11.6 | -0.266 | -- | -- | -- | -- | 0.104 | -- | -- | -- | -- | -- |
| 34.83 | 0.276 | 11.6 | -- | 0.306 | -- | -- | -- | 0.127 | 2.88 | -- | -- | -- | -- |
| 31.88 | 0.263 | 11.6 | -- | -- | 0.512 | -- | -- | 0.04 | -- | 19.98 | 1131 | -- | -- |
| 40.25 | 0.296 | 11.6 | -- | -- | -- | -- | -- | 0.133 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance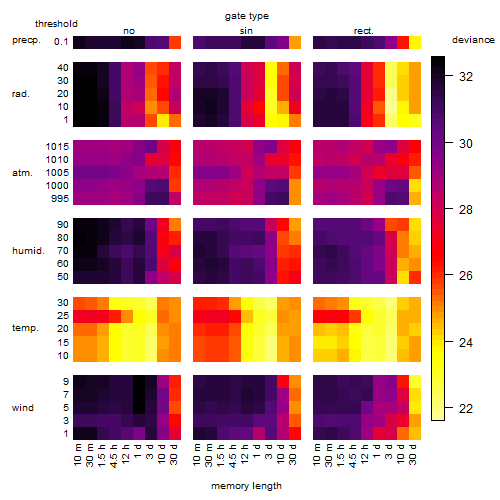
|
Histogram 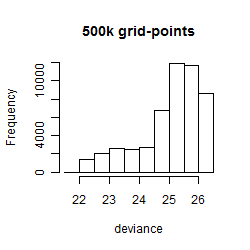
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21.60 | radiation | 10 | 4320 | > th | dose dependent | rect. | 16 | 15 |
| 3 | 21.82 | radiation | 10 | 4320 | > th | dose dependent | rect. | 16 | 2 |
| 15 | 21.84 | radiation | 20 | 4320 | < th | dose independent | rect. | 15 | 16 |
| 17 | 21.98 | temperature | 15 | 4320 | > th | dose dependent | rect. | 21 | 3 |
| 18 | 21.99 | temperature | 15 | 4320 | > th | dose dependent | rect. | 21 | 1 |
| 20 | 22.01 | temperature | 15 | 4320 | > th | dose dependent | rect. | 13 | 13 |
| 28 | 22.03 | temperature | 15 | 4320 | > th | dose dependent | sin | 16 | NA |
| 29 | 22.03 | temperature | 30 | 4320 | < th | dose dependent | rect. | 21 | 1 |
| 32 | 22.03 | temperature | 30 | 4320 | < th | dose dependent | rect. | 17 | 7 |
| 33 | 22.03 | temperature | 15 | 4320 | > th | dose dependent | rect. | 17 | 9 |
| 40 | 22.04 | temperature | 30 | 4320 | < th | dose dependent | rect. | 21 | 3 |
| 98 | 22.08 | temperature | 15 | 4320 | > th | dose dependent | rect. | 23 | 1 |
| 207 | 22.14 | temperature | 30 | 4320 | < th | dose dependent | sin | 15 | NA |
| 249 | 22.16 | radiation | 20 | 4320 | < th | dose independent | rect. | 15 | 3 |
| 320 | 22.18 | temperature | 15 | 4320 | > th | dose dependent | rect. | 17 | 22 |
| 326 | 22.19 | temperature | 15 | 4320 | > th | dose dependent | no | NA | NA |
| 357 | 22.19 | temperature | 15 | 4320 | > th | dose dependent | rect. | 3 | 23 |
| 361 | 22.19 | temperature | 15 | 4320 | > th | dose dependent | rect. | 20 | 23 |
| 393 | 22.20 | temperature | 15 | 4320 | > th | dose dependent | rect. | 1 | 23 |
| 398 | 22.20 | temperature | 15 | 4320 | > th | dose dependent | rect. | 23 | 23 |
| 401 | 22.21 | temperature | 30 | 4320 | < th | dose dependent | rect. | 17 | 2 |
| 812 | 22.34 | radiation | 40 | 4320 | < th | dose dependent | rect. | 16 | 3 |
| 820 | 22.34 | radiation | 30 | 4320 | < th | dose dependent | rect. | 16 | 14 |
| 960 | 22.38 | temperature | 30 | 4320 | < th | dose dependent | no | NA | NA |
| 982 | 22.38 | temperature | 30 | 4320 | < th | dose dependent | rect. | 3 | 23 |