FiT-DB /
Search/
Help/
Sample detail
|
Os07g0680000
|
blastp(Os)/
blastp(At)/
coex///
RAP/
RiceXPro/
SALAD/
ATTED-II
|
|
Description : Similar to Vacuolar sorting receptor 1 precursor (AtVSR1) (Epidermal growth factor receptor-like protein 1) (AtELP1) (AtELP) (BP80-like protein b) (AtBP80b) (Spot 3 protein).
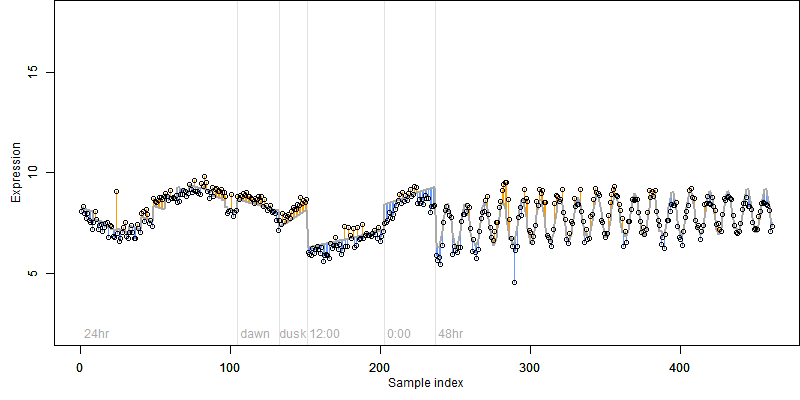

|
log2(Expression) ~
Norm(μ, σ2)
μ = α
+ β1D
+ β2C
+ β3E
+ β4D*C
+ β5D*E
+ γ1N8
| par. |
|
value |
(S.E.) |
|
|
|
|
|
|
| α |
: |
7.75 |
(0.0244) |
R2 |
: |
0.782 |
|
|
|
| β1 |
: |
0.876 |
(0.0893) |
R2dD |
: |
0.0477 |
R2D |
: |
0.0277 |
| β2 |
: |
2.2 |
(0.0581) |
R2dC |
: |
0.689 |
R2C |
: |
0.687 |
| β3 |
: |
-- |
(--) |
R2dE |
: |
-- |
R2E |
: |
-- |
| β4 |
: |
-- |
(--) |
R2dD*C |
: |
-- |
R2D*C |
: |
-- |
| β5 |
: |
-- |
(--) |
R2dD*E |
: |
-- |
R2D*E |
: |
-- |
| γ1 |
: |
0.527 |
(0.0521) |
R2dN8 |
: |
0.0656 |
R2N8 |
: |
0.0375 |
| σ |
: |
0.459 |
|
|
|
|
|
|
|
| deviance |
: |
95.96 |
|
|
|
|
|
|
|
| __ |
|
|
|
|
|
|
|
|
|
|
_____
|
| C |
|
|
| peak time of C |
: |
0.893 |
| E |
|
|
| wheather |
: |
-- |
| threshold |
: |
-- |
| memory length |
: |
-- |
| response mode |
: |
-- |
| dose dependency |
: |
-- |
| G |
|
|
| type of G |
: |
-- |
| peak or start time of G |
: |
-- |
| open length of G |
: |
-- |
|
 __
__
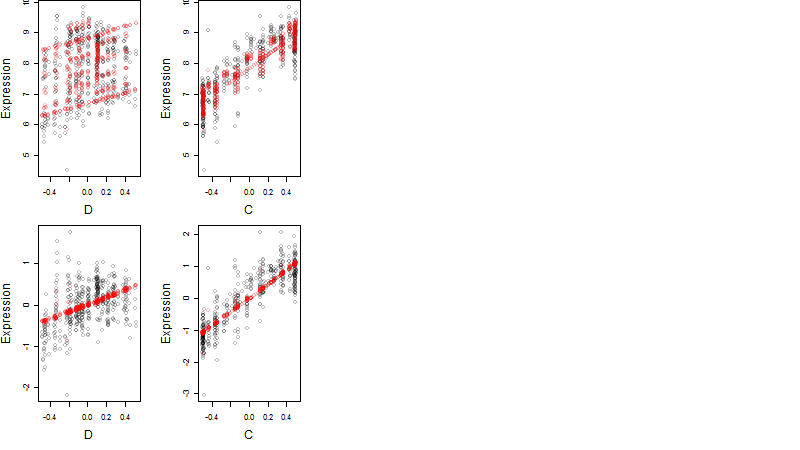
Dependence on each variable
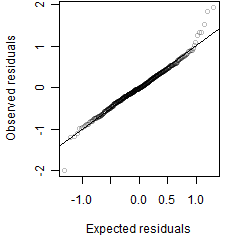
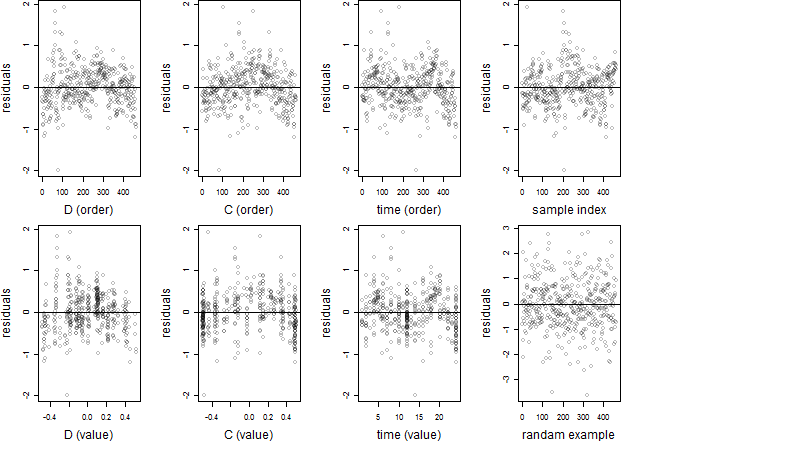
Residual plot
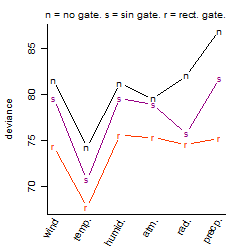
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
| 73.5 |
0.403 |
7.83 |
0.309 |
2.19 |
0.805 |
-0.787 |
-0.878 |
0.284 |
0.898 |
20.4 |
1473 |
-- |
-- |
| 75.29 |
0.404 |
7.81 |
0.346 |
2.18 |
0.932 |
-0.829 |
-- |
0.288 |
0.902 |
20.3 |
2953 |
-- |
-- |
| 75.79 |
0.405 |
7.82 |
0.332 |
2.17 |
0.791 |
-- |
-0.908 |
0.295 |
0.858 |
20.5 |
1381 |
-- |
-- |
| 77.86 |
0.411 |
7.81 |
0.347 |
2.18 |
0.957 |
-- |
-- |
0.278 |
0.858 |
20.15 |
2880 |
-- |
-- |
| 358.18 |
0.881 |
7.81 |
0.176 |
-- |
1.2 |
-- |
-0.748 |
0.23 |
-- |
20.3 |
2459 |
-- |
-- |
| 93.11 |
0.452 |
7.75 |
0.845 |
2.2 |
-- |
-0.828 |
-- |
0.522 |
0.935 |
-- |
-- |
-- |
-- |
| 95.96 |
0.459 |
7.75 |
0.876 |
2.2 |
-- |
-- |
-- |
0.527 |
0.893 |
-- |
-- |
-- |
-- |
| 307.01 |
0.816 |
7.88 |
0.305 |
-- |
1.53 |
-- |
-- |
0.151 |
-- |
25.2 |
941 |
-- |
-- |
| 80.18 |
0.417 |
7.82 |
-- |
2.17 |
1.16 |
-- |
-- |
0.25 |
0.808 |
20 |
2968 |
-- |
-- |
| 398.8 |
0.933 |
7.72 |
0.958 |
-- |
-- |
-- |
-- |
0.568 |
-- |
-- |
-- |
-- |
-- |
| 116.19 |
0.504 |
7.77 |
-- |
2.22 |
-- |
-- |
-- |
0.429 |
0.91 |
-- |
-- |
-- |
-- |
| 306.42 |
0.815 |
7.87 |
-- |
-- |
1.75 |
-- |
-- |
-0.0957 |
-- |
25.5 |
983 |
-- |
-- |
| 422.99 |
0.96 |
7.74 |
-- |
-- |
-- |
-- |
-- |
0.462 |
-- |
-- |
-- |
-- |
-- |
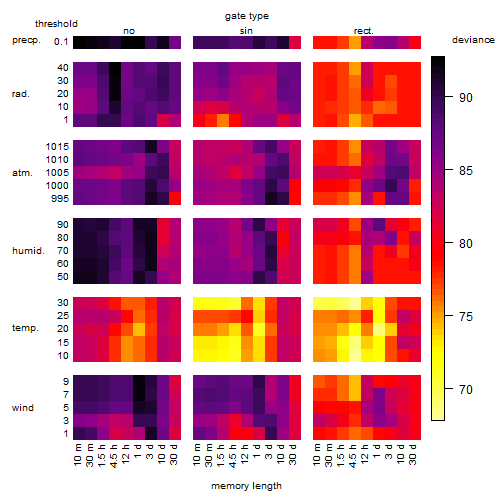
Results of the grid search
Summarized heatmap of deviance
|
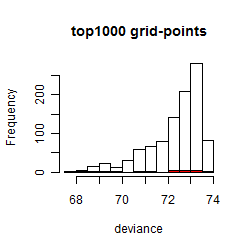
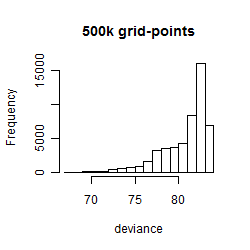
Histogram
|
Local optima within top1000 grid-points
| rank |
deviance |
wheather |
threshold |
memory length |
response mode |
dose dependency |
type of G |
peak or start time of G |
open length of G |
| 1 |
67.79 |
temperature |
30 |
270 |
< th |
dose dependent |
rect. |
10 |
14 |
| 3 |
68.11 |
temperature |
10 |
270 |
> th |
dose dependent |
rect. |
14 |
14 |
| 11 |
68.74 |
temperature |
20 |
1440 |
< th |
dose independent |
rect. |
24 |
4 |
| 12 |
68.77 |
temperature |
20 |
1440 |
> th |
dose independent |
rect. |
1 |
3 |
| 20 |
68.99 |
temperature |
20 |
1440 |
> th |
dose independent |
rect. |
3 |
1 |
| 21 |
69.01 |
temperature |
20 |
1440 |
< th |
dose independent |
rect. |
3 |
1 |
| 47 |
69.77 |
temperature |
30 |
10 |
< th |
dose dependent |
rect. |
12 |
14 |
| 107 |
70.74 |
temperature |
10 |
270 |
> th |
dose dependent |
rect. |
23 |
22 |
| 108 |
70.75 |
temperature |
30 |
270 |
< th |
dose dependent |
sin |
18 |
NA |
| 139 |
70.99 |
temperature |
30 |
270 |
< th |
dose dependent |
rect. |
17 |
8 |
| 173 |
71.26 |
temperature |
20 |
1440 |
> th |
dose independent |
sin |
9 |
NA |
| 174 |
71.26 |
temperature |
10 |
270 |
> th |
dose dependent |
sin |
17 |
NA |
| 201 |
71.45 |
temperature |
20 |
1440 |
< th |
dose independent |
sin |
9 |
NA |
| 304 |
72.11 |
temperature |
25 |
1440 |
< th |
dose dependent |
rect. |
23 |
4 |
| 312 |
72.15 |
temperature |
25 |
1440 |
< th |
dose dependent |
rect. |
2 |
1 |
| 336 |
72.24 |
temperature |
30 |
10 |
< th |
dose dependent |
rect. |
10 |
16 |
| 351 |
72.29 |
temperature |
15 |
1440 |
> th |
dose dependent |
rect. |
2 |
1 |
| 378 |
72.34 |
temperature |
15 |
1440 |
> th |
dose dependent |
rect. |
23 |
4 |
| 457 |
72.58 |
temperature |
30 |
10 |
< th |
dose dependent |
rect. |
20 |
6 |
| 550 |
72.80 |
temperature |
10 |
720 |
> th |
dose dependent |
rect. |
16 |
19 |
| 596 |
72.93 |
temperature |
20 |
1440 |
> th |
dose independent |
rect. |
12 |
18 |
| 633 |
73.00 |
temperature |
20 |
1440 |
< th |
dose independent |
rect. |
12 |
18 |
| 643 |
73.02 |
temperature |
10 |
720 |
> th |
dose dependent |
rect. |
7 |
20 |
| 657 |
73.06 |
temperature |
10 |
720 |
> th |
dose dependent |
rect. |
7 |
14 |
| 680 |
73.10 |
temperature |
30 |
270 |
< th |
dose dependent |
rect. |
21 |
2 |
| 688 |
73.11 |
temperature |
15 |
270 |
> th |
dose dependent |
rect. |
14 |
6 |
| 940 |
73.54 |
temperature |
10 |
1440 |
> th |
dose dependent |
sin |
10 |
NA |
| 986 |
73.61 |
temperature |
10 |
720 |
> th |
dose dependent |
rect. |
8 |
11 |
| 993 |
73.62 |
temperature |
30 |
1440 |
< th |
dose dependent |
sin |
10 |
NA |



 __
__





