Os07g0677300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Peroxidase.
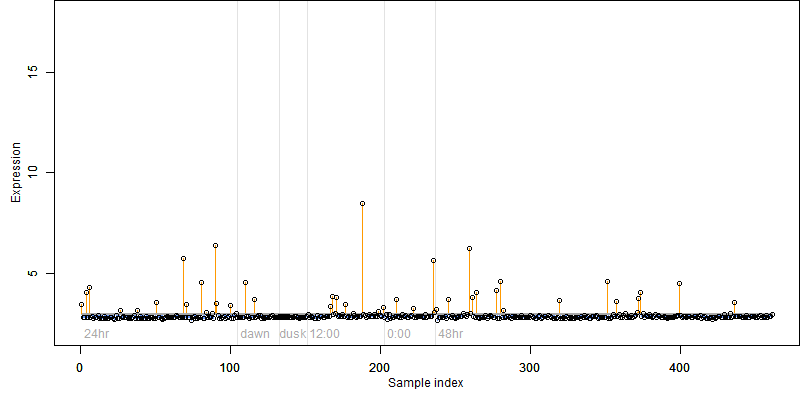

FiT-DB / Search/ Help/ Sample detail
|
Os07g0677300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Peroxidase.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
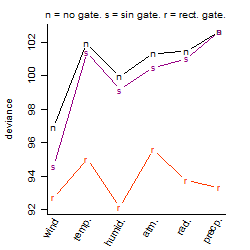
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 96.55 | 0.462 | 2.94 | 0.0647 | 0.147 | -2.33 | -0.191 | -7.97 | 0.0158 | 6.02 | 8.89 | 88 | -- | -- |
| 103.21 | 0.477 | 2.91 | -0.00214 | 0.137 | 0.0797 | -0.175 | -- | 0.0122 | 6.07 | 8.89 | 88 | -- | -- |
| 96.39 | 0.457 | 2.94 | 0.0615 | 0.148 | -2.11 | -- | -7.07 | 0.019 | 5.94 | 8.883 | 76 | -- | -- |
| 102.81 | 0.472 | 2.91 | 0.0235 | 0.145 | -0.34 | -- | -- | 0.0206 | 6.38 | 8.958 | 74 | -- | -- |
| 97.49 | 0.46 | 2.94 | 0.0569 | -- | -2.04 | -- | -7.04 | 0.0187 | -- | 8.869 | 76 | -- | -- |
| 103.23 | 0.476 | 2.91 | 0.00305 | 0.137 | -- | -0.183 | -- | 0.0136 | 6 | -- | -- | -- | -- |
| 103.29 | 0.476 | 2.91 | 0.00197 | 0.137 | -- | -- | -- | 0.0146 | 6.48 | -- | -- | -- | -- |
| 103.32 | 0.475 | 2.91 | 0.0391 | -- | -0.375 | -- | -- | 0.0311 | -- | 8.3 | 127 | -- | -- |
| 102.24 | 0.471 | 2.91 | -- | 0.151 | -0.473 | -- | -- | 0.0243 | 5.96 | 9.389 | 74 | -- | -- |
| 104.27 | 0.477 | 2.91 | 0.00541 | -- | -- | -- | -- | 0.0174 | -- | -- | -- | -- | -- |
| 103.29 | 0.475 | 2.91 | -- | 0.137 | -- | -- | -- | 0.0144 | 6.48 | -- | -- | -- | -- |
| 102.64 | 0.473 | 2.91 | -- | -- | -0.58 | -- | -- | 0.0282 | -- | 8.3 | 76 | -- | -- |
| 104.27 | 0.477 | 2.91 | -- | -- | -- | -- | -- | 0.0168 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram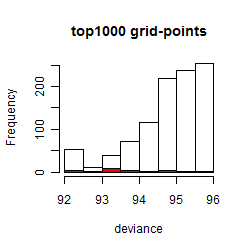 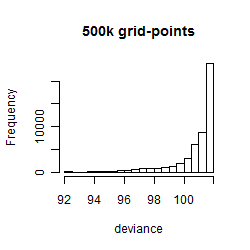
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 92.16 | humidity | 90.0 | 10 | < th | dose dependent | rect. | 4 | 2 |
| 3 | 92.24 | humidity | 80.0 | 10 | < th | dose independent | rect. | 1 | 5 |
| 7 | 92.25 | humidity | 80.0 | 10 | < th | dose dependent | rect. | 1 | 5 |
| 55 | 92.74 | wind | 5.0 | 720 | > th | dose dependent | rect. | 4 | 1 |
| 56 | 92.75 | wind | 5.0 | 720 | > th | dose independent | rect. | 4 | 1 |
| 69 | 93.26 | wind | 5.0 | 90 | > th | dose dependent | rect. | 8 | 4 |
| 70 | 93.30 | wind | 9.0 | 90 | > th | dose dependent | rect. | 10 | 1 |
| 71 | 93.31 | precipitation | 0.1 | 1440 | > th | dose independent | rect. | 15 | 1 |
| 72 | 93.31 | precipitation | 0.1 | 1440 | > th | dose dependent | rect. | 15 | 1 |
| 74 | 93.32 | wind | 5.0 | 90 | > th | dose dependent | rect. | 10 | 1 |
| 79 | 93.36 | precipitation | 0.1 | 1440 | < th | dose independent | rect. | 15 | 1 |
| 80 | 93.36 | precipitation | 0.1 | 1440 | < th | dose dependent | rect. | 15 | 1 |
| 81 | 93.37 | wind | 9.0 | 90 | > th | dose independent | rect. | 10 | 1 |
| 131 | 93.76 | radiation | 1.0 | 90 | > th | dose independent | rect. | 4 | 1 |
| 141 | 93.76 | radiation | 1.0 | 90 | > th | dose dependent | rect. | 4 | 1 |
| 167 | 93.92 | wind | 5.0 | 10 | > th | dose dependent | rect. | 11 | 1 |
| 177 | 94.01 | humidity | 90.0 | 1440 | > th | dose dependent | rect. | 15 | 2 |
| 218 | 94.26 | wind | 9.0 | 10 | > th | dose independent | rect. | 11 | 1 |
| 340 | 94.60 | wind | 5.0 | 90 | > th | dose dependent | sin | 5 | NA |
| 414 | 94.82 | wind | 7.0 | 90 | > th | dose independent | sin | 5 | NA |
| 482 | 94.94 | wind | 7.0 | 90 | < th | dose independent | sin | 6 | NA |
| 518 | 95.03 | temperature | 20.0 | 90 | < th | dose dependent | rect. | 4 | 1 |
| 783 | 95.58 | atmosphere | 1005.0 | 10 | > th | dose dependent | rect. | 5 | 1 |
| 921 | 95.84 | wind | 1.0 | 1440 | > th | dose independent | rect. | 17 | 1 |