Os07g0646000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.
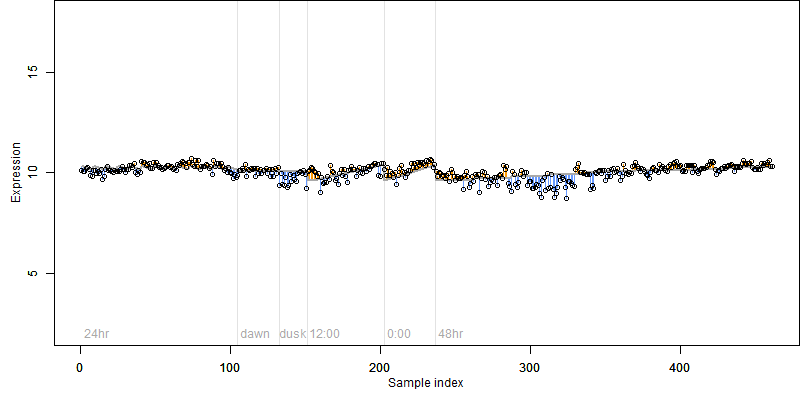

FiT-DB / Search/ Help/ Sample detail
|
Os07g0646000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
Dependence on each variable

Residual plot

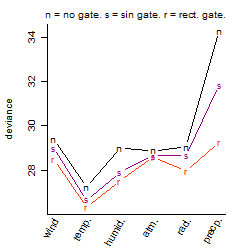
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 26.97 | 0.244 | 9.9 | 1.34 | 0.238 | 0.37 | 0.269 | -1.2 | 0.585 | 2.17 | 22.9 | 1643 | -- | -- |
| 30.12 | 0.256 | 9.94 | 0.887 | 0.265 | 0.343 | 0.36 | -- | 0.483 | 2.19 | 22.85 | 1569 | -- | -- |
| 27.18 | 0.243 | 9.9 | 1.34 | 0.237 | 0.377 | -- | -1.19 | 0.581 | 2.04 | 22.88 | 1571 | -- | -- |
| 30.3 | 0.256 | 9.95 | 0.89 | 0.249 | 0.348 | -- | -- | 0.46 | 2.19 | 22.8 | 1569 | -- | -- |
| 30.62 | 0.258 | 9.9 | 1.33 | -- | 0.364 | -- | -1.18 | 0.579 | -- | 22.84 | 1626 | -- | -- |
| 35.46 | 0.279 | 9.98 | 0.772 | 0.234 | -- | 0.274 | -- | 0.333 | 2.26 | -- | -- | -- | -- |
| 35.73 | 0.28 | 9.98 | 0.764 | 0.234 | -- | -- | -- | 0.331 | 2.14 | -- | -- | -- | -- |
| 33.8 | 0.271 | 9.94 | 0.902 | -- | 0.351 | -- | -- | 0.47 | -- | 22.88 | 1627 | -- | -- |
| 49.94 | 0.329 | 9.97 | -- | 0.283 | 0.146 | -- | -- | 0.301 | 1.98 | 22.82 | 960 | -- | -- |
| 39 | 0.292 | 9.97 | 0.774 | -- | -- | -- | -- | 0.337 | -- | -- | -- | -- | -- |
| 51.12 | 0.334 | 10 | -- | 0.248 | -- | -- | -- | 0.246 | 2.21 | -- | -- | -- | -- |
| 51.46 | 0.335 | 9.98 | -- | -- | 0.178 | -- | -- | 0.299 | -- | 25.8 | 25 | -- | -- |
| 54.79 | 0.345 | 9.99 | -- | -- | -- | -- | -- | 0.251 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram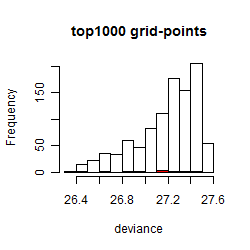 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 26.32 | temperature | 15 | 270 | > th | dose dependent | rect. | 12 | 22 |
| 2 | 26.43 | temperature | 20 | 1440 | > th | dose dependent | rect. | 13 | 10 |
| 43 | 26.63 | temperature | 25 | 1440 | > th | dose dependent | rect. | 13 | 5 |
| 52 | 26.65 | temperature | 20 | 1440 | > th | dose dependent | sin | 16 | NA |
| 121 | 26.83 | temperature | 15 | 270 | > th | dose dependent | rect. | 3 | 17 |
| 175 | 26.93 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 23 |
| 205 | 26.99 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 21 |
| 360 | 27.17 | temperature | 15 | 270 | > th | dose dependent | rect. | 24 | 21 |
| 378 | 27.19 | temperature | 30 | 1440 | < th | dose dependent | rect. | 13 | 18 |
| 380 | 27.19 | temperature | 20 | 1440 | > th | dose dependent | rect. | 2 | 20 |
| 395 | 27.20 | temperature | 30 | 1440 | < th | dose dependent | rect. | 13 | 15 |
| 425 | 27.22 | temperature | 20 | 1440 | > th | dose dependent | no | NA | NA |
| 605 | 27.32 | temperature | 30 | 1440 | < th | dose dependent | sin | 15 | NA |
| 754 | 27.41 | temperature | 30 | 270 | < th | dose dependent | rect. | 7 | 21 |
| 918 | 27.50 | humidity | 90 | 14400 | > th | dose dependent | rect. | 21 | 6 |