Os07g0642200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Lipolytic enzyme, G-D-S-L family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os07g0642200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Lipolytic enzyme, G-D-S-L family protein.
|
|
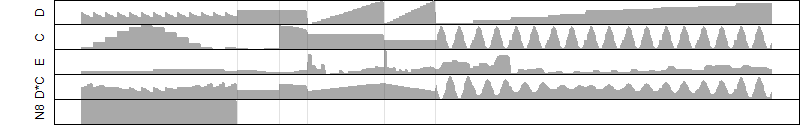
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
Dependence on each variable

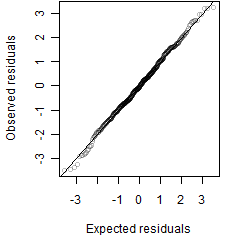
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = radiation,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 696.05 | 1.24 | 6.48 | -3 | 0.555 | -6.68 | -3.11 | -4.72 | -0.552 | 16.4 | 0.6532 | 1463 | -- | -- |
| 704.55 | 1.24 | 6.55 | -3.36 | 0.436 | -5.69 | -3.45 | -- | -0.604 | 16.9 | 0.8108 | 1475 | -- | -- |
| 717.84 | 1.25 | 6.51 | -3.23 | 0.69 | -6.52 | -- | -3.75 | -0.615 | 15 | 0.6563 | 1454 | -- | -- |
| 722.08 | 1.25 | 6.54 | -3.3 | 0.743 | -5.4 | -- | -- | -0.619 | 13.9 | 1.322 | 1445 | -- | -- |
| 735.17 | 1.26 | 6.5 | -3.16 | -- | -6.62 | -- | -3.68 | -0.601 | -- | 0.6554 | 1477 | -- | -- |
| 1029.88 | 1.5 | 6.39 | -2.04 | 0.811 | -- | -2.82 | -- | 0.0811 | 15.1 | -- | -- | -- | -- |
| 1049.64 | 1.52 | 6.39 | -2.09 | 0.836 | -- | -- | -- | 0.0618 | 13.5 | -- | -- | -- | -- |
| 733.85 | 1.26 | 6.58 | -3.16 | -- | -5.51 | -- | -- | -0.807 | -- | 1.83 | 2986 | -- | -- |
| 979.98 | 1.46 | 6.46 | -- | 0.449 | -4.09 | -- | -- | -0.151 | 15.1 | 1.657 | 1486 | -- | -- |
| 1092.43 | 1.54 | 6.4 | -2.13 | -- | -- | -- | -- | 0.0436 | -- | -- | -- | -- | -- |
| 1165.29 | 1.6 | 6.34 | -- | 0.874 | -- | -- | -- | 0.295 | 13.6 | -- | -- | -- | -- |
| 968.74 | 1.45 | 6.51 | -- | -- | -5.03 | -- | -- | -0.423 | -- | 2.562 | 3031 | -- | -- |
| 1211.9 | 1.62 | 6.35 | -- | -- | -- | -- | -- | 0.28 | -- | -- | -- | -- | -- |
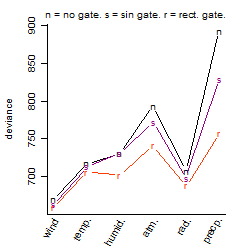
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 659.81 | wind | 5 | 43200 | > th | dose dependent | rect. | 11 | 18 |
| 12 | 663.04 | wind | 5 | 43200 | > th | dose independent | rect. | 3 | 19 |
| 15 | 663.54 | wind | 5 | 43200 | > th | dose dependent | sin | 17 | NA |
| 17 | 663.62 | wind | 5 | 43200 | > th | dose independent | rect. | 10 | 20 |
| 20 | 663.77 | wind | 5 | 43200 | > th | dose independent | rect. | 8 | 14 |
| 31 | 664.52 | wind | 5 | 43200 | > th | dose independent | rect. | 24 | 21 |
| 46 | 665.09 | wind | 5 | 43200 | > th | dose dependent | rect. | 12 | 11 |
| 54 | 665.31 | wind | 5 | 43200 | > th | dose dependent | rect. | 19 | 2 |
| 57 | 665.47 | wind | 5 | 43200 | < th | dose independent | rect. | 3 | 19 |
| 58 | 665.54 | wind | 7 | 43200 | < th | dose independent | rect. | 12 | 17 |
| 85 | 666.51 | wind | 5 | 43200 | > th | dose independent | rect. | 3 | 22 |
| 91 | 666.63 | wind | 5 | 43200 | < th | dose independent | rect. | 24 | 22 |
| 94 | 666.72 | wind | 5 | 43200 | > th | dose independent | sin | 0 | NA |
| 95 | 666.77 | wind | 5 | 43200 | < th | dose independent | rect. | 9 | 21 |
| 103 | 667.03 | wind | 9 | 43200 | > th | dose dependent | rect. | 19 | 3 |
| 131 | 667.59 | wind | 9 | 43200 | > th | dose independent | rect. | 19 | 3 |
| 136 | 667.74 | wind | 7 | 43200 | > th | dose independent | rect. | 13 | 16 |
| 138 | 667.77 | wind | 5 | 43200 | < th | dose independent | rect. | 7 | 15 |
| 154 | 668.16 | wind | 7 | 43200 | < th | dose independent | sin | 16 | NA |
| 204 | 669.19 | wind | 5 | 43200 | > th | dose independent | rect. | 3 | 15 |
| 231 | 669.57 | wind | 5 | 43200 | < th | dose independent | sin | 0 | NA |
| 232 | 669.59 | wind | 5 | 43200 | > th | dose independent | no | NA | NA |
| 258 | 670.30 | wind | 5 | 43200 | > th | dose independent | rect. | 20 | 23 |
| 259 | 670.33 | wind | 7 | 43200 | > th | dose independent | sin | 16 | NA |
| 266 | 670.46 | wind | 5 | 43200 | < th | dose independent | rect. | 3 | 15 |
| 289 | 670.99 | wind | 5 | 43200 | < th | dose independent | rect. | 24 | 18 |
| 297 | 671.17 | wind | 5 | 43200 | < th | dose independent | no | NA | NA |
| 309 | 671.40 | wind | 5 | 43200 | < th | dose independent | rect. | 20 | 23 |
| 316 | 671.54 | wind | 5 | 43200 | > th | dose independent | rect. | 8 | 8 |
| 351 | 672.33 | wind | 5 | 43200 | > th | dose independent | rect. | 9 | 6 |
| 361 | 672.52 | wind | 7 | 43200 | < th | dose independent | rect. | 17 | 4 |
| 406 | 673.35 | wind | 5 | 43200 | > th | dose independent | rect. | 17 | 23 |
| 411 | 673.47 | wind | 5 | 43200 | < th | dose independent | rect. | 9 | 6 |
| 427 | 673.69 | wind | 5 | 43200 | < th | dose independent | rect. | 8 | 8 |
| 435 | 673.84 | wind | 5 | 43200 | > th | dose dependent | rect. | 3 | 18 |
| 446 | 674.00 | wind | 5 | 43200 | > th | dose dependent | rect. | 24 | 21 |
| 467 | 674.21 | wind | 5 | 43200 | < th | dose independent | rect. | 17 | 23 |
| 506 | 674.75 | wind | 7 | 43200 | > th | dose independent | rect. | 17 | 4 |
| 637 | 676.56 | wind | 7 | 43200 | < th | dose independent | rect. | 14 | 9 |
| 677 | 677.16 | wind | 5 | 43200 | > th | dose dependent | rect. | 17 | 4 |
| 691 | 677.42 | wind | 9 | 43200 | < th | dose independent | rect. | 19 | 2 |
| 708 | 677.62 | wind | 5 | 43200 | > th | dose dependent | rect. | 5 | 21 |
| 807 | 679.37 | wind | 5 | 43200 | > th | dose dependent | no | NA | NA |
| 902 | 682.16 | wind | 5 | 43200 | > th | dose dependent | rect. | 20 | 23 |
| 929 | 683.09 | wind | 5 | 43200 | > th | dose dependent | rect. | 17 | 23 |