Os07g0419300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Thaumatin-like protein.

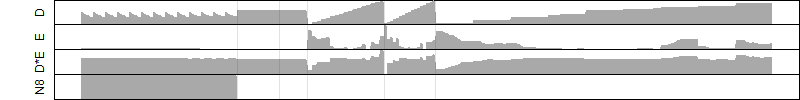
FiT-DB / Search/ Help/ Sample detail
|
Os07g0419300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Thaumatin-like protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
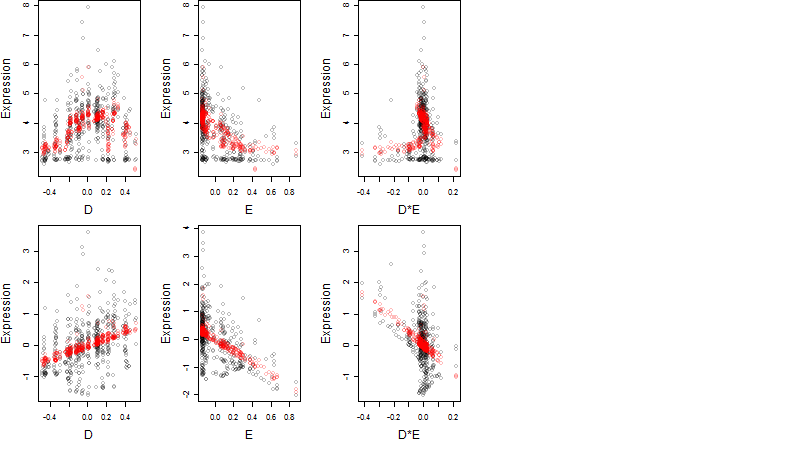
Dependence on each variable
Residual plot

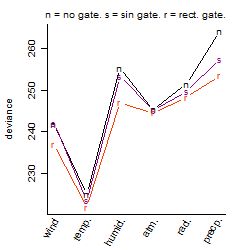
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 222.8 | 0.701 | 3.74 | 1.12 | 0.519 | -2.21 | 0.397 | -4.04 | 0.324 | 10.6 | 24.19 | 837 | -- | -- |
| 241.03 | 0.723 | 3.82 | 0.535 | 0.504 | -1.69 | -0.512 | -- | 0.205 | 10.3 | 25 | 762 | -- | -- |
| 223.34 | 0.696 | 3.74 | 1.1 | 0.515 | -2.21 | -- | -3.83 | 0.317 | 10.6 | 24.1 | 837 | -- | -- |
| 241.85 | 0.724 | 3.82 | 0.507 | 0.514 | -1.71 | -- | -- | 0.185 | 10.2 | 25 | 764 | -- | -- |
| 235.74 | 0.715 | 3.74 | 1.14 | -- | -2.13 | -- | -4.08 | 0.329 | -- | 24.25 | 1356 | -- | -- |
| 284.79 | 0.791 | 3.75 | 0.996 | 0.264 | -- | -0.412 | -- | 0.542 | 9.98 | -- | -- | -- | -- |
| 285.44 | 0.791 | 3.75 | 0.981 | 0.266 | -- | -- | -- | 0.542 | 10.2 | -- | -- | -- | -- |
| 252.96 | 0.741 | 3.83 | 0.454 | -- | -1.66 | -- | -- | 0.191 | -- | 25.1 | 1417 | -- | -- |
| 247.41 | 0.733 | 3.84 | -- | 0.532 | -1.94 | -- | -- | 0.0937 | 10.5 | 24.91 | 820 | -- | -- |
| 289.72 | 0.795 | 3.76 | 0.977 | -- | -- | -- | -- | 0.541 | -- | -- | -- | -- | -- |
| 310.81 | 0.825 | 3.77 | -- | 0.261 | -- | -- | -- | 0.432 | 9.94 | -- | -- | -- | -- |
| 257.97 | 0.748 | 3.86 | -- | -- | -1.89 | -- | -- | 0.0944 | -- | 25.2 | 1420 | -- | -- |
| 314.91 | 0.828 | 3.78 | -- | -- | -- | -- | -- | 0.433 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance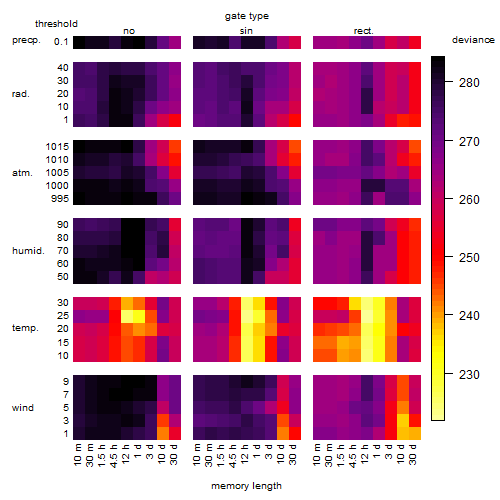
|
Histogram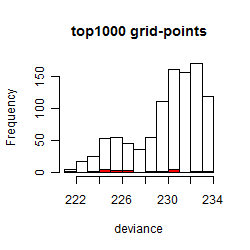 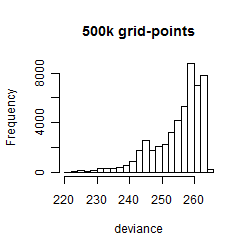
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 221.84 | temperature | 25 | 720 | < th | dose dependent | rect. | 20 | 19 |
| 32 | 223.49 | temperature | 25 | 720 | < th | dose dependent | sin | 8 | NA |
| 76 | 224.68 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 16 |
| 78 | 224.71 | temperature | 25 | 720 | < th | dose dependent | rect. | 24 | 23 |
| 83 | 224.77 | temperature | 25 | 720 | < th | dose dependent | rect. | 13 | 23 |
| 90 | 224.92 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 98 | 225.03 | temperature | 25 | 720 | < th | dose dependent | rect. | 10 | 23 |
| 138 | 225.81 | temperature | 15 | 720 | > th | dose dependent | sin | 8 | NA |
| 152 | 226.03 | temperature | 25 | 720 | < th | dose dependent | rect. | 6 | 23 |
| 164 | 226.39 | temperature | 10 | 720 | > th | dose dependent | rect. | 23 | 8 |
| 180 | 226.67 | temperature | 15 | 720 | > th | dose dependent | rect. | 23 | 12 |
| 239 | 228.21 | temperature | 30 | 720 | < th | dose dependent | rect. | 1 | 8 |
| 321 | 229.41 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 6 |
| 451 | 230.40 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 2 |
| 452 | 230.41 | temperature | 25 | 1440 | < th | dose dependent | rect. | 4 | 1 |
| 514 | 230.78 | temperature | 25 | 1440 | < th | dose dependent | rect. | 24 | 1 |
| 524 | 230.82 | temperature | 15 | 1440 | > th | dose dependent | rect. | 4 | 1 |
| 721 | 232.07 | temperature | 10 | 720 | > th | dose dependent | rect. | 3 | 8 |
| 891 | 233.05 | temperature | 20 | 1440 | > th | dose independent | rect. | 3 | 1 |