Os07g0277900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.


FiT-DB / Search/ Help/ Sample detail
|
Os07g0277900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
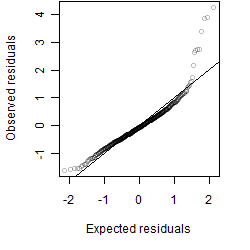
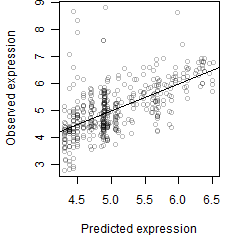 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 242.34 | 0.731 | 5.1 | 0.903 | 0.678 | 1.32 | 0.718 | 0.414 | -0.329 | 9.75 | 22.3 | 1949 | -- | -- |
| 242.69 | 0.726 | 5.08 | 1.05 | 0.687 | 1.33 | 0.775 | -- | -0.293 | 9.69 | 22.2 | 1944 | -- | -- |
| 244 | 0.728 | 5.11 | 0.913 | 0.663 | 1.36 | -- | 0.586 | -0.347 | 9.76 | 22.1 | 1959 | -- | -- |
| 244.76 | 0.729 | 5.08 | 1.09 | 0.67 | 1.34 | -- | -- | -0.295 | 9.82 | 22.1 | 1963 | -- | -- |
| 254.35 | 0.743 | 5.09 | 0.946 | -- | 1.4 | -- | 0.457 | -0.336 | -- | 22.26 | 792 | -- | -- |
| 306.55 | 0.821 | 5.19 | 0.518 | 0.744 | -- | 0.727 | -- | -0.753 | 9.76 | -- | -- | -- | -- |
| 308.54 | 0.823 | 5.19 | 0.546 | 0.739 | -- | -- | -- | -0.752 | 9.84 | -- | -- | -- | -- |
| 254.96 | 0.744 | 5.1 | 0.984 | -- | 1.36 | -- | -- | -0.294 | -- | 22.26 | 800 | -- | -- |
| 271.69 | 0.768 | 5.11 | -- | 0.658 | 1.07 | -- | -- | -0.454 | 9.49 | 22.3 | 1943 | -- | -- |
| 341.21 | 0.863 | 5.21 | 0.538 | -- | -- | -- | -- | -0.751 | -- | -- | -- | -- | -- |
| 316.4 | 0.832 | 5.2 | -- | 0.738 | -- | -- | -- | -0.812 | 9.79 | -- | -- | -- | -- |
| 278.73 | 0.778 | 5.14 | -- | -- | 1.17 | -- | -- | -0.451 | -- | 22.38 | 781 | -- | -- |
| 348.86 | 0.872 | 5.22 | -- | -- | -- | -- | -- | -0.811 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 240.89 | temperature | 20 | 1440 | > th | dose dependent | rect. | 1 | 1 |
| 14 | 243.52 | temperature | 25 | 720 | < th | dose dependent | rect. | 10 | 21 |
| 27 | 244.12 | temperature | 25 | 720 | < th | dose dependent | rect. | 13 | 18 |
| 78 | 245.15 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 86 | 245.26 | temperature | 30 | 1440 | < th | dose dependent | rect. | 1 | 1 |
| 90 | 245.31 | temperature | 25 | 720 | < th | dose dependent | rect. | 24 | 23 |
| 95 | 245.41 | temperature | 25 | 720 | < th | dose dependent | rect. | 3 | 23 |
| 138 | 246.08 | temperature | 25 | 720 | < th | dose dependent | rect. | 21 | 23 |
| 170 | 246.45 | temperature | 25 | 720 | < th | dose dependent | sin | 12 | NA |
| 173 | 246.48 | temperature | 25 | 1440 | < th | dose dependent | sin | 14 | NA |
| 187 | 246.63 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 9 |
| 255 | 247.17 | temperature | 20 | 720 | > th | dose dependent | sin | 10 | NA |
| 357 | 247.85 | temperature | 20 | 720 | > th | dose dependent | rect. | 19 | 14 |
| 370 | 247.93 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 1 |
| 741 | 250.33 | temperature | 15 | 720 | > th | dose dependent | rect. | 15 | 18 |