Os07g0247000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Pectinesterase inhibitor domain containing protein.

FiT-DB / Search/ Help/ Sample detail
|
Os07g0247000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Pectinesterase inhibitor domain containing protein.
|
|
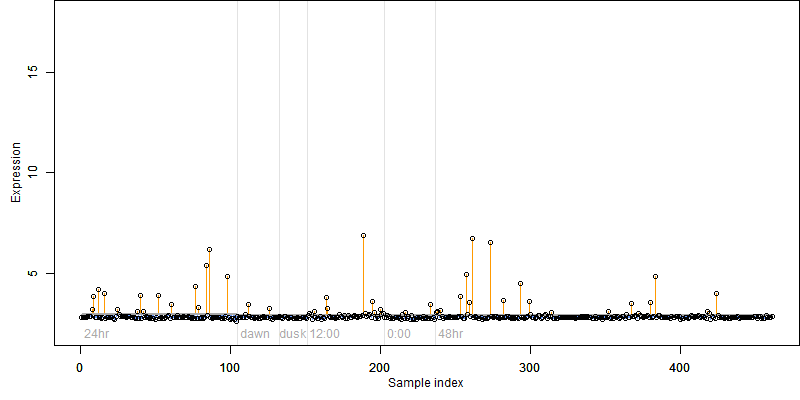
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

Dependence on each variable

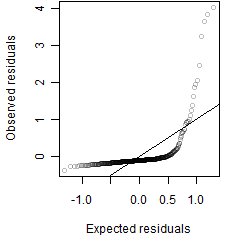
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 88.74 | 0.443 | 2.9 | -0.0862 | 0.145 | -1.77 | -0.483 | -5.6 | 0.0588 | 6.71 | 8.8 | 76 | -- | -- |
| 92.82 | 0.452 | 2.88 | -0.115 | 0.135 | -0.398 | -0.458 | -- | 0.0602 | 6.21 | 8.8 | 76 | -- | -- |
| 89.32 | 0.44 | 2.9 | -0.0954 | 0.144 | -1.72 | -- | -5.56 | 0.0591 | 7.03 | 8.8 | 76 | -- | -- |
| 93.23 | 0.453 | 2.88 | -0.119 | 0.139 | -0.384 | -- | -- | 0.0622 | 7.39 | 8.9 | 79 | -- | -- |
| 90.38 | 0.443 | 2.9 | -0.0974 | -- | -1.69 | -- | -5.61 | 0.0604 | -- | 8.8 | 76 | -- | -- |
| 93.41 | 0.453 | 2.88 | -0.14 | 0.137 | -- | -0.426 | -- | 0.0532 | 6.55 | -- | -- | -- | -- |
| 93.79 | 0.454 | 2.88 | -0.145 | 0.139 | -- | -- | -- | 0.0552 | 7.56 | -- | -- | -- | -- |
| 94.25 | 0.454 | 2.88 | -0.117 | -- | -0.387 | -- | -- | 0.0645 | -- | 8.9 | 79 | -- | -- |
| 93.59 | 0.453 | 2.87 | -- | 0.138 | -0.437 | -- | -- | 0.0761 | 7.42 | 8.846 | 76 | -- | -- |
| 94.82 | 0.455 | 2.88 | -0.143 | -- | -- | -- | -- | 0.0573 | -- | -- | -- | -- | -- |
| 94.34 | 0.454 | 2.87 | -- | 0.138 | -- | -- | -- | 0.0713 | 7.63 | -- | -- | -- | -- |
| 94.6 | 0.454 | 2.88 | -- | -- | -0.439 | -- | -- | 0.0781 | -- | 8.866 | 76 | -- | -- |
| 95.36 | 0.456 | 2.88 | -- | -- | -- | -- | -- | 0.0731 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance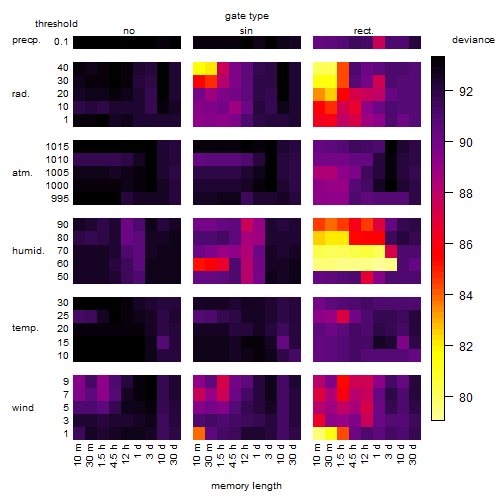
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 79.06 | humidity | 60 | 1440 | > th | dose independent | rect. | 4 | 5 |
| 2 | 79.28 | humidity | 60 | 1440 | > th | dose independent | rect. | 23 | 10 |
| 3 | 79.30 | humidity | 60 | 270 | < th | dose independent | rect. | 8 | 1 |
| 29 | 79.30 | humidity | 60 | 270 | < th | dose dependent | rect. | 8 | 1 |
| 108 | 79.50 | humidity | 60 | 90 | < th | dose independent | rect. | 18 | 15 |
| 109 | 79.51 | humidity | 60 | 90 | < th | dose dependent | rect. | 18 | 15 |
| 112 | 79.53 | humidity | 60 | 1440 | > th | dose independent | rect. | 20 | 13 |
| 113 | 79.53 | humidity | 60 | 30 | < th | dose independent | rect. | 17 | 17 |
| 130 | 79.56 | humidity | 60 | 30 | < th | dose independent | rect. | 9 | 1 |
| 163 | 79.56 | humidity | 60 | 10 | < th | dose dependent | rect. | 9 | 1 |
| 319 | 79.61 | wind | 1 | 10 | < th | dose dependent | rect. | 9 | 1 |
| 355 | 79.79 | wind | 1 | 10 | < th | dose independent | rect. | 9 | 1 |
| 362 | 79.97 | radiation | 40 | 30 | > th | dose dependent | rect. | 9 | 1 |
| 587 | 81.70 | radiation | 40 | 10 | > th | dose dependent | sin | 12 | NA |
| 725 | 82.37 | radiation | 40 | 10 | > th | dose independent | rect. | 9 | 1 |
| 930 | 83.79 | wind | 1 | 10 | < th | dose independent | sin | 21 | NA |
| 948 | 83.91 | humidity | 90 | 30 | < th | dose dependent | rect. | 24 | 10 |
| 963 | 84.02 | wind | 1 | 10 | < th | dose dependent | sin | 21 | NA |
| 984 | 84.08 | radiation | 40 | 10 | > th | dose independent | sin | 11 | NA |