Os07g0188800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Methylmalonate-semialdehyde dehydrogenase (EC 1.2.1.27).
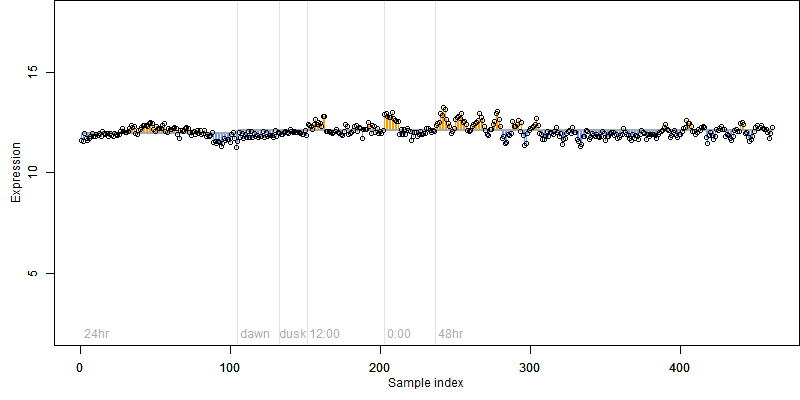

FiT-DB / Search/ Help/ Sample detail
|
Os07g0188800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Methylmalonate-semialdehyde dehydrogenase (EC 1.2.1.27).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
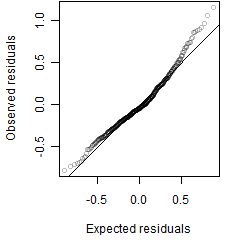
 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16.37 | 0.19 | 12 | 0.0715 | 0.451 | -0.565 | -1.03 | 1.28 | 0.0558 | 18.4 | 19.73 | 3897 | -- | -- |
| 17.41 | 0.194 | 12.1 | -0.239 | 0.432 | -0.578 | -1.12 | -- | -0.03 | 18.1 | 17.57 | 3961 | -- | -- |
| 18.81 | 0.202 | 12 | 0.086 | 0.445 | -0.545 | -- | 1.25 | 0.0494 | 17.9 | 19.09 | 4111 | -- | -- |
| 19.62 | 0.206 | 12.1 | -0.221 | 0.441 | -0.639 | -- | -- | -0.0338 | 18 | 14.68 | 3940 | -- | -- |
| 28.78 | 0.25 | 12 | -0.00154 | -- | -0.572 | -- | 0.912 | 0.0179 | -- | 18.2 | 4172 | -- | -- |
| 24.73 | 0.233 | 12.1 | -0.515 | 0.462 | -- | -0.992 | -- | -0.154 | 18 | -- | -- | -- | -- |
| 26.99 | 0.243 | 12.1 | -0.504 | 0.459 | -- | -- | -- | -0.157 | 17.6 | -- | -- | -- | -- |
| 29.25 | 0.252 | 12.1 | -0.254 | -- | -0.648 | -- | -- | -0.0337 | -- | 14.2 | 2751 | -- | -- |
| 20.59 | 0.211 | 12.1 | -- | 0.429 | -0.743 | -- | -- | 0.00538 | 18 | 12.96 | 4140 | -- | -- |
| 37.95 | 0.288 | 12.1 | -0.519 | -- | -- | -- | -- | -0.168 | -- | -- | -- | -- | -- |
| 33.69 | 0.272 | 12.1 | -- | 0.468 | -- | -- | -- | -0.101 | 17.6 | -- | -- | -- | -- |
| 30.33 | 0.257 | 12.1 | -- | -- | -0.778 | -- | -- | 0.00419 | -- | 14.4 | 4179 | -- | -- |
| 45.05 | 0.313 | 12.1 | -- | -- | -- | -- | -- | -0.11 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram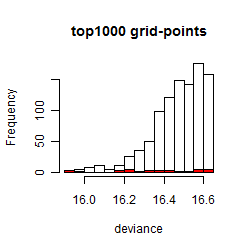 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 15.91 | temperature | 20 | 4320 | < th | dose independent | rect. | 21 | 3 |
| 3 | 15.93 | temperature | 20 | 4320 | > th | dose independent | rect. | 21 | 3 |
| 5 | 15.98 | temperature | 20 | 4320 | < th | dose independent | rect. | 19 | 6 |
| 37 | 16.19 | temperature | 25 | 1440 | < th | dose dependent | rect. | 3 | 1 |
| 40 | 16.20 | temperature | 15 | 1440 | > th | dose dependent | rect. | 3 | 1 |
| 43 | 16.21 | temperature | 10 | 10 | > th | dose dependent | sin | 15 | NA |
| 44 | 16.21 | temperature | 15 | 4320 | > th | dose dependent | rect. | 21 | 1 |
| 46 | 16.21 | temperature | 15 | 4320 | > th | dose dependent | rect. | 20 | 4 |
| 51 | 16.22 | temperature | 15 | 4320 | > th | dose dependent | rect. | 20 | 8 |
| 78 | 16.27 | temperature | 15 | 4320 | > th | dose dependent | rect. | 23 | 1 |
| 110 | 16.32 | temperature | 25 | 4320 | < th | dose dependent | rect. | 21 | 1 |
| 131 | 16.34 | temperature | 10 | 90 | > th | dose dependent | sin | 16 | NA |
| 161 | 16.36 | temperature | 30 | 4320 | < th | dose dependent | rect. | 20 | 4 |
| 169 | 16.36 | temperature | 30 | 4320 | < th | dose dependent | rect. | 20 | 8 |
| 185 | 16.37 | temperature | 20 | 4320 | > th | dose dependent | rect. | 15 | 9 |
| 256 | 16.41 | temperature | 20 | 4320 | > th | dose dependent | sin | 15 | NA |
| 352 | 16.45 | temperature | 30 | 4320 | < th | dose dependent | rect. | 23 | 1 |
| 430 | 16.48 | temperature | 25 | 4320 | < th | dose independent | rect. | 11 | 18 |
| 635 | 16.55 | temperature | 20 | 4320 | > th | dose independent | sin | 14 | NA |
| 665 | 16.56 | temperature | 20 | 4320 | < th | dose independent | sin | 14 | NA |
| 671 | 16.56 | temperature | 25 | 4320 | > th | dose independent | rect. | 11 | 18 |
| 677 | 16.56 | temperature | 30 | 4320 | < th | dose dependent | sin | 13 | NA |
| 706 | 16.56 | temperature | 15 | 270 | > th | dose dependent | rect. | 9 | 22 |
| 944 | 16.63 | temperature | 10 | 720 | > th | dose dependent | rect. | 10 | 19 |
| 962 | 16.64 | temperature | 20 | 4320 | > th | dose dependent | no | NA | NA |
| 987 | 16.64 | temperature | 20 | 4320 | > th | dose dependent | rect. | 1 | 23 |
| 990 | 16.64 | temperature | 20 | 4320 | > th | dose dependent | rect. | 17 | 23 |
| 999 | 16.65 | temperature | 25 | 4320 | < th | dose independent | rect. | 19 | 21 |