Os07g0162900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Esterase/lipase/thioesterase domain containing protein.

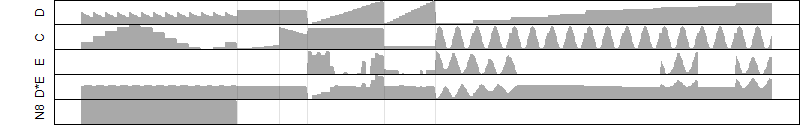
FiT-DB / Search/ Help/ Sample detail
|
Os07g0162900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Esterase/lipase/thioesterase domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
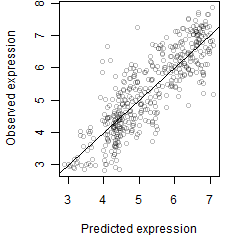 __
__

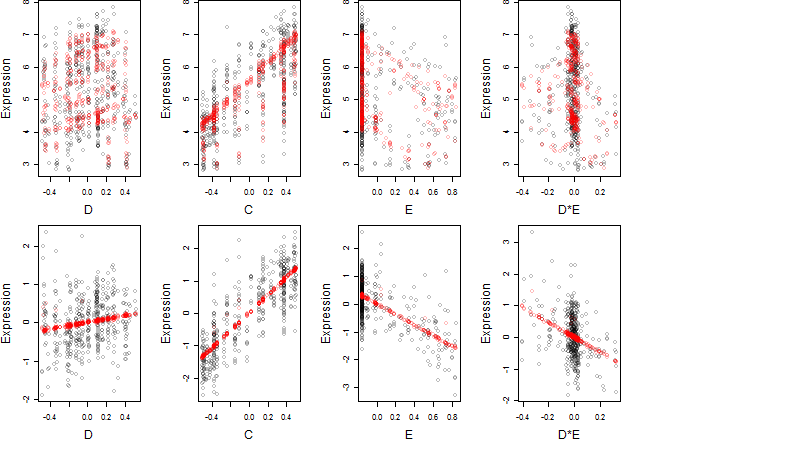
Dependence on each variable
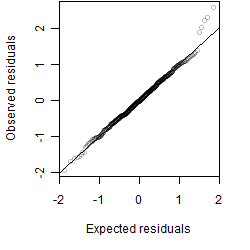
Residual plot
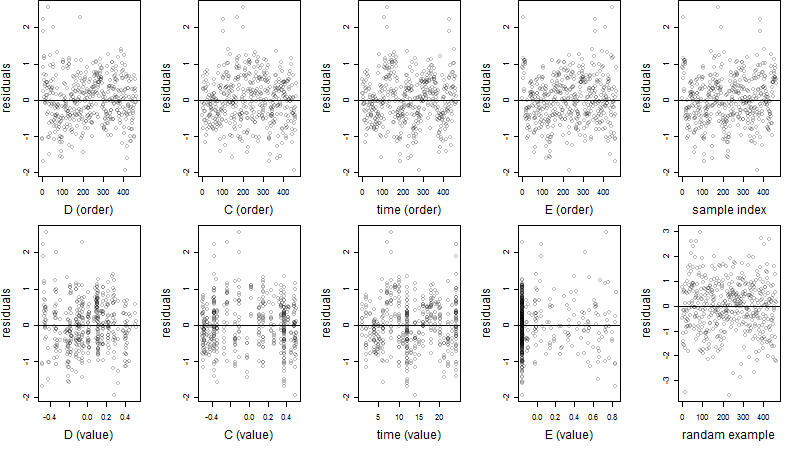
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 196.4 | 0.658 | 5.24 | 0.509 | 2.77 | -1.84 | 0.449 | -2.54 | 0.106 | 14.9 | 21.6 | 709 | 0.9 | -- |
| 212.12 | 0.678 | 5.28 | -0.0504 | 2.76 | -1.87 | -0.43 | -- | 0.0143 | 14.8 | 21.7 | 638 | 0.462 | -- |
| 196.21 | 0.652 | 5.23 | 0.442 | 2.76 | -1.89 | -- | -2.31 | 0.112 | 14.8 | 21.7 | 652 | 0.482 | -- |
| 212.66 | 0.679 | 5.29 | -0.0657 | 2.77 | -1.87 | -- | -- | -0.0214 | 14.8 | 21.66 | 632 | 0.422 | -- |
| 527.57 | 1.07 | 5.27 | -0.0161 | -- | -2.22 | -- | -1.15 | -0.0965 | -- | 22.5 | 103 | 1.35 | -- |
| 305.87 | 0.82 | 5.21 | 0.383 | 2.59 | -- | 0.212 | -- | 0.396 | 15.6 | -- | -- | -- | -- |
| 306.01 | 0.819 | 5.21 | 0.387 | 2.59 | -- | -- | -- | 0.397 | 15.6 | -- | -- | -- | -- |
| 271.68 | 0.768 | 5.21 | 0.258 | -- | -2.43 | -- | -- | 0.323 | -- | 27.52 | 3 | 2.3 | -- |
| 212.76 | 0.679 | 5.29 | -- | 2.76 | -1.85 | -- | -- | -0.0109 | 14.8 | 21.66 | 632 | 0.447 | -- |
| 680.51 | 1.22 | 5.21 | 0.278 | -- | -- | -- | -- | 0.328 | -- | -- | -- | -- | -- |
| 309.95 | 0.824 | 5.22 | -- | 2.59 | -- | -- | -- | 0.354 | 15.6 | -- | -- | -- | -- |
| 273.4 | 0.77 | 5.22 | -- | -- | -2.43 | -- | -- | 0.288 | -- | 27.5 | 4 | 2.29 | -- |
| 682.55 | 1.22 | 5.21 | -- | -- | -- | -- | -- | 0.297 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance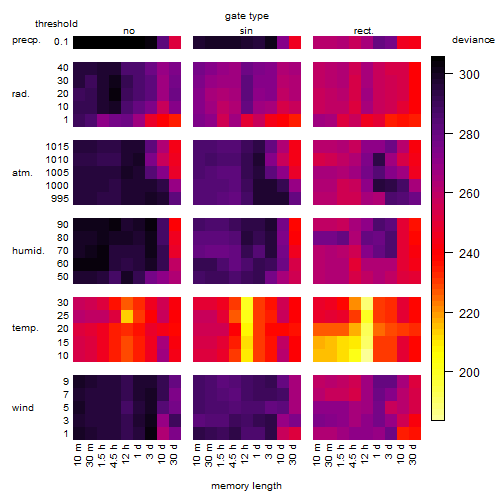
|
Histogram 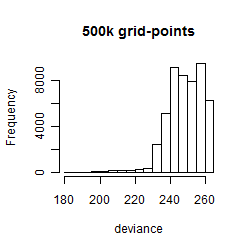
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 183.67 | temperature | 10 | 720 | > th | dose dependent | rect. | 21 | 12 |
| 3 | 187.55 | temperature | 10 | 720 | > th | dose dependent | rect. | 4 | 5 |
| 4 | 187.76 | temperature | 30 | 720 | < th | dose dependent | rect. | 1 | 4 |
| 18 | 193.55 | temperature | 30 | 720 | < th | dose dependent | rect. | 24 | 11 |
| 73 | 200.22 | temperature | 30 | 720 | < th | dose dependent | sin | 7 | NA |
| 103 | 201.98 | temperature | 10 | 270 | > th | dose dependent | rect. | 4 | 13 |
| 192 | 205.94 | temperature | 15 | 720 | > th | dose dependent | rect. | 21 | 19 |
| 253 | 208.06 | temperature | 20 | 720 | > th | dose dependent | sin | 7 | NA |
| 266 | 208.54 | temperature | 10 | 720 | > th | dose dependent | sin | 7 | NA |
| 436 | 213.26 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 502 | 215.73 | temperature | 25 | 720 | < th | dose dependent | rect. | 5 | 23 |
| 547 | 216.86 | temperature | 10 | 720 | > th | dose dependent | rect. | 13 | 20 |
| 738 | 222.28 | temperature | 20 | 720 | > th | dose independent | rect. | 4 | 5 |
| 752 | 222.82 | temperature | 10 | 270 | > th | dose dependent | rect. | 2 | 23 |