Os07g0142600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.


FiT-DB / Search/ Help/ Sample detail
|
Os07g0142600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

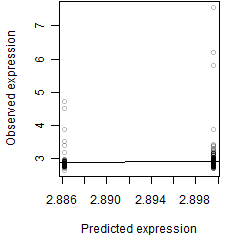 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = < th, dose dependency = dose dependent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 32.28 | 0.267 | 2.95 | -0.254 | 0.0829 | 19.5 | -0.0579 | -91.1 | 0.00575 | 8.98 | 999.5 | 89 | 11.5 | 0.513 |
| 41.09 | 0.299 | 2.93 | -0.11 | 0.0242 | 6.75 | 0.995 | -- | -0.0476 | 6.81 | 996.9 | 142 | 10 | 1.61 |
| 32.29 | 0.265 | 2.95 | -0.255 | 0.0831 | 19.5 | -- | -90.8 | 0.00579 | 9.15 | 999.5 | 90 | 11.5 | 0.512 |
| 32.31 | 0.266 | 2.89 | 0.0622 | 0.0822 | 4.62 | -- | -- | 0.00603 | 9.13 | 997.4 | 40 | 11.8 | 1.51 |
| 32.69 | 0.268 | 2.95 | -0.252 | -- | 19.5 | -- | -91.1 | 0.00639 | -- | 999.5 | 90 | 11.7 | 0.85 |
| 53.48 | 0.343 | 2.89 | 0.0858 | 0.11 | -- | 0.0496 | -- | -0.0037 | 10.2 | -- | -- | -- | -- |
| 53.49 | 0.343 | 2.89 | 0.0876 | 0.11 | -- | -- | -- | -0.00368 | 10 | -- | -- | -- | -- |
| 32.7 | 0.267 | 2.9 | 0.0618 | -- | 4.65 | -- | -- | 0.00653 | -- | 997.4 | 42 | 11.8 | 1.48 |
| 32.41 | 0.267 | 2.89 | -- | 0.0822 | 4.63 | -- | -- | -0.000874 | 9.07 | 997.4 | 40 | 11.8 | 1.51 |
| 54.22 | 0.344 | 2.9 | 0.0862 | -- | -- | -- | -- | -0.00378 | -- | -- | -- | -- | -- |
| 53.69 | 0.343 | 2.9 | -- | 0.109 | -- | -- | -- | -0.0134 | 9.98 | -- | -- | -- | -- |
| 32.8 | 0.268 | 2.9 | -- | -- | 4.66 | -- | -- | -0.000303 | -- | 997.4 | 42 | 11.8 | 1.48 |
| 54.41 | 0.344 | 2.9 | -- | -- | -- | -- | -- | -0.0133 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance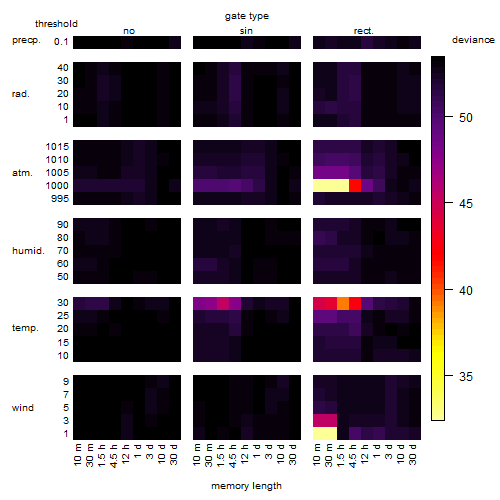
|
Histogram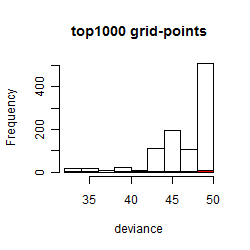 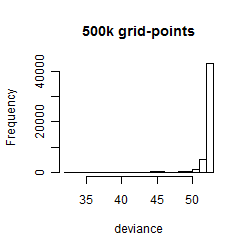
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 32.38 | atmosphere | 1000 | 90 | < th | dose dependent | rect. | 10 | 1 |
| 3 | 32.39 | atmosphere | 1000 | 10 | < th | dose dependent | rect. | 11 | 1 |
| 13 | 32.59 | wind | 1 | 10 | < th | dose independent | rect. | 12 | 1 |
| 15 | 32.59 | wind | 1 | 10 | < th | dose dependent | rect. | 12 | 1 |
| 18 | 34.34 | atmosphere | 1000 | 90 | < th | dose independent | rect. | 10 | 1 |
| 20 | 34.68 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 11 | 1 |
| 63 | 39.23 | temperature | 30 | 90 | > th | dose dependent | rect. | 10 | 1 |
| 73 | 41.68 | atmosphere | 1000 | 270 | < th | dose independent | rect. | 8 | 4 |
| 94 | 42.55 | temperature | 30 | 270 | > th | dose dependent | rect. | 8 | 3 |
| 130 | 42.97 | temperature | 30 | 270 | > th | dose independent | rect. | 8 | 1 |
| 165 | 43.54 | temperature | 30 | 90 | > th | dose independent | rect. | 10 | 1 |
| 300 | 45.46 | temperature | 30 | 90 | > th | dose dependent | sin | 9 | NA |
| 414 | 46.85 | atmosphere | 1000 | 10 | < th | dose dependent | rect. | 6 | 7 |
| 457 | 47.61 | temperature | 30 | 270 | > th | dose independent | sin | 9 | NA |
| 528 | 48.42 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 6 | 7 |
| 551 | 48.55 | temperature | 30 | 270 | < th | dose independent | sin | 9 | NA |
| 578 | 48.84 | atmosphere | 1000 | 720 | < th | dose independent | rect. | 24 | 12 |
| 659 | 49.25 | temperature | 25 | 30 | > th | dose independent | rect. | 7 | 1 |
| 698 | 49.43 | atmosphere | 1000 | 10 | < th | dose dependent | rect. | 2 | 11 |
| 970 | 49.84 | atmosphere | 1000 | 270 | > th | dose independent | sin | 2 | NA |
| 972 | 49.84 | atmosphere | 1000 | 270 | < th | dose independent | sin | 2 | NA |