Os07g0124900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Allergen V5/Tpx-1 related family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os07g0124900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Allergen V5/Tpx-1 related family protein.
|
|
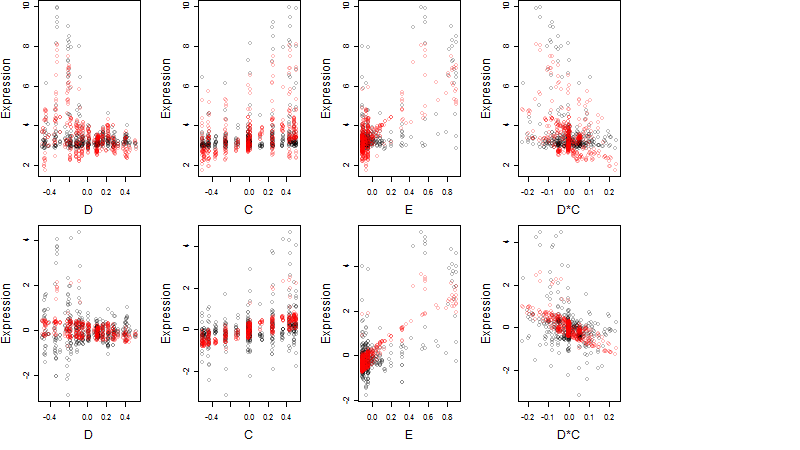
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
Dependence on each variable
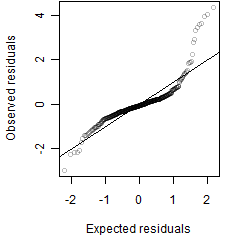
Residual plot

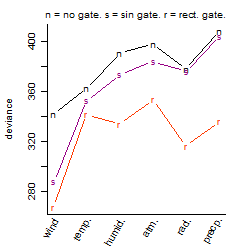
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose independent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 259.78 | 0.757 | 3.37 | -0.639 | 0.773 | 1.65 | -3.37 | -7.44 | -0.0298 | 5.76 | 7.2 | 2914 | 7.59 | -- |
| 268.8 | 0.764 | 3.46 | -0.303 | 0.822 | 3 | -3.92 | -- | 0.0068 | 5.96 | 7.1 | 2537 | 9.33 | -- |
| 285.78 | 0.787 | 3.35 | -0.588 | 0.765 | 1.78 | -- | -7.02 | 0.00846 | 6.03 | 7.031 | 2913 | 8.36 | -- |
| 295.87 | 0.801 | 3.46 | -0.246 | 0.781 | 3.24 | -- | -- | -0.0374 | 6.37 | 7.046 | 2547 | 9.54 | -- |
| 315.42 | 0.827 | 3.36 | -0.557 | -- | 1.85 | -- | -7.02 | 0.0293 | -- | 7.1 | 2829 | 8.16 | -- |
| 444.21 | 0.988 | 3.56 | -1.24 | 0.849 | -- | -3.88 | -- | -0.445 | 5.29 | -- | -- | -- | -- |
| 481.66 | 1.03 | 3.55 | -1.25 | 0.826 | -- | -- | -- | -0.427 | 5.45 | -- | -- | -- | -- |
| 325.84 | 0.841 | 3.46 | -0.239 | -- | 3.26 | -- | -- | 0.00389 | -- | 7.1 | 2600 | 8.96 | -- |
| 297.49 | 0.803 | 3.46 | -- | 0.769 | 3.34 | -- | -- | -0.0183 | 6.49 | 7.053 | 2547 | 9.51 | -- |
| 517.18 | 1.06 | 3.56 | -1.22 | -- | -- | -- | -- | -0.407 | -- | -- | -- | -- | -- |
| 522.74 | 1.07 | 3.52 | -- | 0.805 | -- | -- | -- | -0.288 | 5.51 | -- | -- | -- | -- |
| 327.19 | 0.842 | 3.47 | -- | -- | 3.35 | -- | -- | 0.0193 | -- | 6.968 | 2600 | 9.11 | -- |
| 556.49 | 1.1 | 3.53 | -- | -- | -- | -- | -- | -0.272 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 267.53 | wind | 5 | 4320 | < th | dose independent | rect. | 24 | 1 |
| 2 | 268.42 | wind | 5 | 4320 | > th | dose independent | rect. | 24 | 1 |
| 3 | 274.77 | wind | 7 | 4320 | > th | dose independent | rect. | 17 | 4 |
| 4 | 275.25 | wind | 7 | 4320 | > th | dose dependent | rect. | 17 | 4 |
| 9 | 277.33 | wind | 9 | 4320 | > th | dose independent | rect. | 17 | 11 |
| 10 | 277.41 | wind | 9 | 4320 | > th | dose independent | rect. | 17 | 9 |
| 12 | 277.51 | wind | 7 | 4320 | > th | dose independent | rect. | 16 | 13 |
| 17 | 277.93 | wind | 7 | 4320 | > th | dose independent | rect. | 15 | 18 |
| 23 | 278.22 | wind | 7 | 4320 | < th | dose independent | rect. | 17 | 4 |
| 32 | 278.85 | wind | 7 | 4320 | < th | dose independent | rect. | 16 | 13 |
| 36 | 278.95 | wind | 7 | 4320 | < th | dose independent | rect. | 16 | 16 |
| 47 | 279.55 | wind | 7 | 4320 | < th | dose independent | rect. | 15 | 18 |
| 64 | 280.42 | wind | 7 | 4320 | > th | dose dependent | rect. | 17 | 1 |
| 67 | 280.61 | wind | 9 | 4320 | < th | dose independent | rect. | 17 | 1 |
| 72 | 280.89 | wind | 9 | 4320 | < th | dose independent | rect. | 17 | 9 |
| 76 | 281.34 | wind | 7 | 4320 | > th | dose dependent | rect. | 17 | 11 |
| 202 | 288.02 | wind | 7 | 4320 | > th | dose independent | sin | 13 | NA |
| 219 | 289.34 | wind | 7 | 4320 | < th | dose independent | sin | 13 | NA |
| 304 | 295.21 | wind | 7 | 4320 | > th | dose dependent | sin | 12 | NA |
| 320 | 296.38 | wind | 5 | 4320 | > th | dose dependent | rect. | 24 | 1 |
| 684 | 316.42 | radiation | 1 | 1440 | < th | dose independent | rect. | 11 | 1 |
| 685 | 316.42 | radiation | 1 | 1440 | < th | dose dependent | rect. | 11 | 1 |
| 738 | 318.45 | wind | 5 | 4320 | > th | dose independent | rect. | 21 | 1 |
| 740 | 318.52 | wind | 5 | 4320 | < th | dose independent | rect. | 21 | 1 |
| 817 | 321.60 | wind | 9 | 4320 | > th | dose dependent | rect. | 22 | 1 |
| 829 | 321.96 | wind | 9 | 4320 | > th | dose independent | rect. | 22 | 1 |
| 887 | 323.88 | wind | 1 | 720 | < th | dose independent | rect. | 23 | 1 |