Os07g0124700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to PLETHORA1.


FiT-DB / Search/ Help/ Sample detail
|
Os07g0124700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to PLETHORA1.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
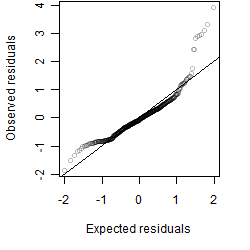
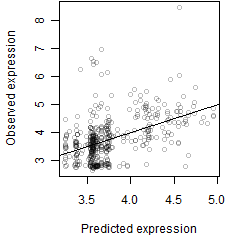
Dependence on each variable

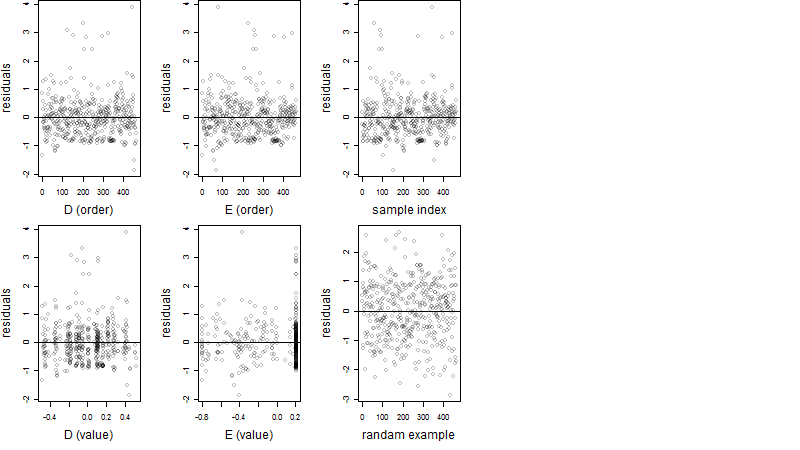
Residual plot
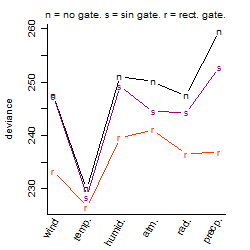
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 222.35 | 0.701 | 3.73 | 0.823 | 0.0561 | -1.3 | 0.392 | -0.71 | 0.197 | 1.34 | 21.4 | 914 | -- | -- |
| 223.64 | 0.697 | 3.69 | 0.978 | 0.0631 | -1.21 | 0.331 | -- | 0.276 | 1.72 | 21.82 | 1022 | -- | -- |
| 222.83 | 0.695 | 3.73 | 0.835 | 0.0536 | -1.29 | -- | -0.619 | 0.211 | 1.24 | 21.45 | 956 | -- | -- |
| 224.05 | 0.697 | 3.7 | 0.971 | 0.0534 | -1.19 | -- | -- | 0.263 | 2.37 | 21.8 | 1022 | -- | -- |
| 223 | 0.696 | 3.73 | 0.833 | -- | -1.27 | -- | -0.613 | 0.207 | -- | 21.45 | 956 | -- | -- |
| 275.86 | 0.779 | 3.78 | 0.527 | 0.149 | -- | -0.241 | -- | -0.108 | 11.5 | -- | -- | -- | -- |
| 276.03 | 0.778 | 3.78 | 0.516 | 0.156 | -- | -- | -- | -0.11 | 10.5 | -- | -- | -- | -- |
| 224.19 | 0.697 | 3.7 | 0.987 | -- | -1.2 | -- | -- | 0.258 | -- | 21.78 | 1015 | -- | -- |
| 244.86 | 0.729 | 3.74 | -- | 0.0495 | -0.934 | -- | -- | 0.115 | 2.23 | 22.2 | 1040 | -- | -- |
| 277.52 | 0.778 | 3.79 | 0.513 | -- | -- | -- | -- | -0.111 | -- | -- | -- | -- | -- |
| 283.04 | 0.787 | 3.8 | -- | 0.153 | -- | -- | -- | -0.168 | 10.3 | -- | -- | -- | -- |
| 244.99 | 0.729 | 3.74 | -- | -- | -0.925 | -- | -- | 0.111 | -- | 22.28 | 1040 | -- | -- |
| 284.47 | 0.787 | 3.8 | -- | -- | -- | -- | -- | -0.168 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 226.52 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 5 |
| 2 | 226.54 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 1 |
| 40 | 228.32 | temperature | 25 | 720 | < th | dose dependent | rect. | 24 | 22 |
| 42 | 228.36 | temperature | 25 | 1440 | < th | dose dependent | sin | 15 | NA |
| 99 | 229.39 | temperature | 25 | 720 | < th | dose dependent | rect. | 17 | 20 |
| 118 | 229.54 | temperature | 10 | 720 | > th | dose dependent | sin | 8 | NA |
| 137 | 229.69 | temperature | 25 | 720 | < th | dose dependent | rect. | 19 | 23 |
| 151 | 229.77 | temperature | 15 | 720 | > th | dose dependent | rect. | 20 | 18 |
| 166 | 229.82 | temperature | 25 | 720 | < th | dose dependent | rect. | 11 | 23 |
| 185 | 229.96 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 193 | 230.03 | temperature | 30 | 720 | < th | dose dependent | sin | 8 | NA |
| 195 | 230.05 | temperature | 15 | 720 | > th | dose dependent | rect. | 20 | 14 |
| 257 | 230.54 | temperature | 25 | 720 | < th | dose dependent | rect. | 4 | 23 |
| 293 | 230.75 | temperature | 25 | 720 | < th | dose dependent | rect. | 6 | 23 |
| 348 | 231.12 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 12 |
| 514 | 231.83 | temperature | 10 | 1440 | > th | dose dependent | rect. | 20 | 2 |
| 614 | 232.20 | temperature | 10 | 1440 | > th | dose dependent | sin | 12 | NA |
| 811 | 232.85 | temperature | 20 | 720 | > th | dose independent | rect. | 20 | 22 |
| 858 | 232.99 | temperature | 20 | 720 | > th | dose independent | rect. | 20 | 19 |
| 867 | 233.01 | temperature | 20 | 720 | > th | dose independent | sin | 8 | NA |
| 876 | 233.03 | temperature | 25 | 1440 | < th | dose dependent | rect. | 4 | 1 |
| 916 | 233.13 | temperature | 20 | 720 | > th | dose independent | rect. | 12 | 21 |
| 917 | 233.13 | temperature | 20 | 720 | < th | dose independent | rect. | 1 | 21 |
| 968 | 233.27 | temperature | 20 | 720 | < th | dose independent | rect. | 3 | 19 |
| 980 | 233.30 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 4 |
| 988 | 233.32 | wind | 9 | 43200 | > th | dose dependent | rect. | 18 | 12 |