Os06g0693300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.


FiT-DB / Search/ Help/ Sample detail
|
Os06g0693300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
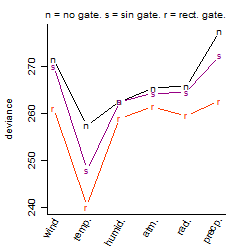
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 236.35 | 0.722 | 5.11 | 1.18 | 1.14 | -0.459 | -0.381 | 4.76 | 0.224 | 17.2 | 20.18 | 4683 | 6.02 | 0.957 |
| 278.15 | 0.777 | 5.27 | 1.03 | 1.1 | -1.3 | -0.269 | -- | 0.164 | 17.3 | 17.5 | 23086 | 6.48 | 1.32 |
| 236.71 | 0.717 | 5.11 | 1.18 | 1.14 | -0.463 | -- | 4.77 | 0.224 | 17.1 | 20.16 | 4680 | 6.01 | 0.955 |
| 269.29 | 0.764 | 5.27 | 0.941 | 1.18 | -1.61 | -- | -- | 0.139 | 17.3 | 18.4 | 21741 | 6.52 | 1.42 |
| 303.97 | 0.812 | 5.1 | 1.1 | -- | -0.515 | -- | 4.83 | 0.208 | -- | 19.93 | 4682 | 5.98 | 0.8 |
| 293.27 | 0.803 | 5.37 | -0.499 | 1.19 | -- | -0.588 | -- | -0.343 | 17.3 | -- | -- | -- | -- |
| 294.09 | 0.803 | 5.37 | -0.497 | 1.18 | -- | -- | -- | -0.346 | 17.2 | -- | -- | -- | -- |
| 333.22 | 0.854 | 5.33 | -0.543 | -- | -1.05 | -- | -- | -0.267 | -- | 18.4 | 21780 | 6.52 | 1.48 |
| 273.62 | 0.77 | 5.31 | -- | 1.08 | -1.19 | -- | -- | -0.0347 | 17.2 | 17.2 | 21743 | 6.58 | 1.4 |
| 367.07 | 0.895 | 5.35 | -0.538 | -- | -- | -- | -- | -0.375 | -- | -- | -- | -- | -- |
| 300.61 | 0.811 | 5.36 | -- | 1.19 | -- | -- | -- | -0.291 | 17.2 | -- | -- | -- | -- |
| 341.01 | 0.863 | 5.32 | -- | -- | -1.05 | -- | -- | -0.207 | -- | 18.4 | 21780 | 6.52 | 1.48 |
| 374.72 | 0.904 | 5.34 | -- | -- | -- | -- | -- | -0.316 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance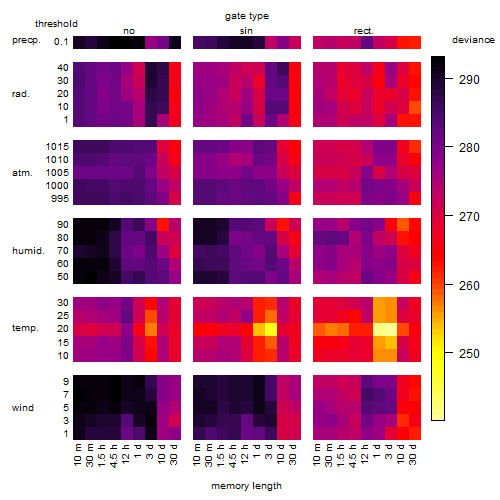
|
Histogram 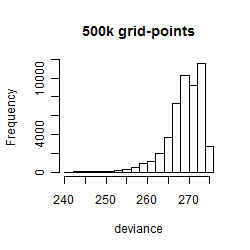
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 240.09 | temperature | 20 | 4320 | > th | dose independent | rect. | 6 | 1 |
| 2 | 240.45 | temperature | 20 | 4320 | < th | dose independent | rect. | 5 | 2 |
| 5 | 241.69 | temperature | 20 | 1440 | > th | dose independent | rect. | 3 | 3 |
| 6 | 242.11 | temperature | 20 | 1440 | < th | dose independent | rect. | 3 | 3 |
| 7 | 242.15 | temperature | 20 | 4320 | < th | dose independent | rect. | 2 | 5 |
| 11 | 242.66 | temperature | 20 | 4320 | > th | dose independent | rect. | 3 | 1 |
| 17 | 243.10 | temperature | 20 | 4320 | < th | dose independent | rect. | 3 | 1 |
| 26 | 243.46 | temperature | 20 | 4320 | < th | dose independent | rect. | 22 | 9 |
| 32 | 243.64 | temperature | 20 | 4320 | > th | dose independent | rect. | 22 | 9 |
| 127 | 246.48 | temperature | 20 | 4320 | < th | dose independent | rect. | 23 | 1 |
| 153 | 247.44 | temperature | 20 | 4320 | > th | dose independent | rect. | 23 | 1 |
| 172 | 247.93 | temperature | 20 | 4320 | > th | dose independent | sin | 9 | NA |
| 195 | 248.38 | temperature | 20 | 4320 | < th | dose independent | sin | 9 | NA |
| 271 | 250.30 | temperature | 20 | 1440 | > th | dose dependent | rect. | 5 | 2 |
| 405 | 252.87 | temperature | 20 | 4320 | > th | dose dependent | rect. | 22 | 2 |
| 424 | 253.16 | temperature | 20 | 1440 | > th | dose dependent | rect. | 24 | 8 |
| 471 | 253.73 | temperature | 20 | 4320 | > th | dose dependent | rect. | 20 | 12 |
| 543 | 254.47 | temperature | 20 | 4320 | > th | dose dependent | sin | 11 | NA |
| 723 | 255.52 | temperature | 30 | 1440 | < th | dose dependent | rect. | 2 | 1 |
| 860 | 256.21 | temperature | 25 | 4320 | > th | dose dependent | rect. | 16 | 1 |
| 923 | 256.52 | temperature | 30 | 4320 | < th | dose dependent | rect. | 6 | 1 |
| 970 | 256.72 | temperature | 30 | 1440 | < th | dose dependent | rect. | 24 | 4 |