Os06g0642500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Cytochrome P450 71A1 (EC 1.14.-.-) (CYPLXXIA1) (ARP-2).

FiT-DB / Search/ Help/ Sample detail
|
Os06g0642500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Cytochrome P450 71A1 (EC 1.14.-.-) (CYPLXXIA1) (ARP-2).
|
|
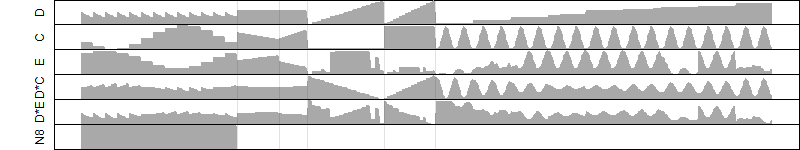
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
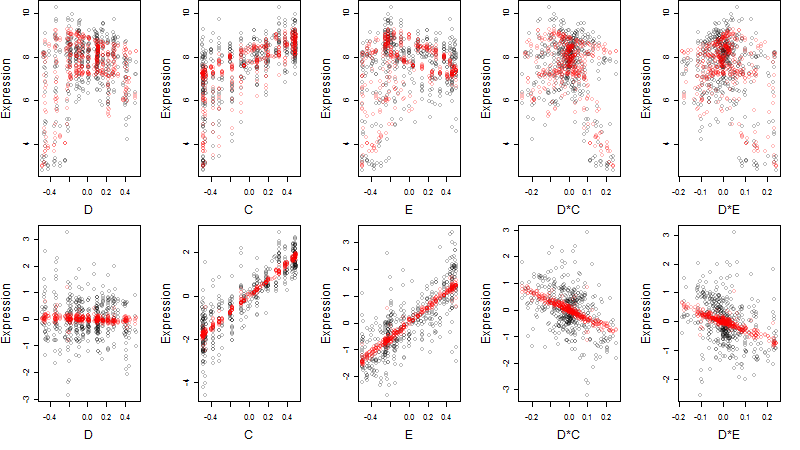
Dependence on each variable
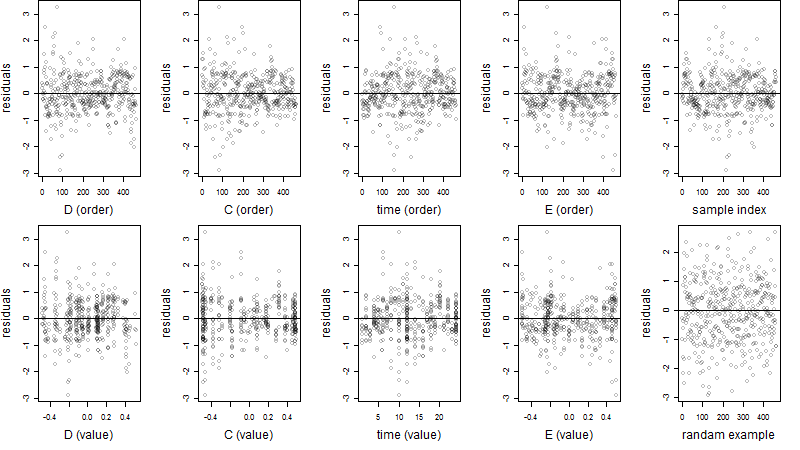
Residual plot
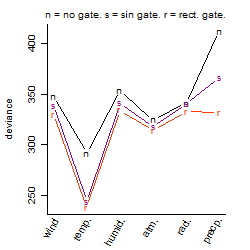
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 237.07 | 0.723 | 7.71 | -0.103 | 3.66 | 2.84 | -3.31 | -3.06 | -0.0262 | 23.4 | 20.9 | 745 | 1.96 | -- |
| 252.86 | 0.741 | 7.66 | 0.569 | 3.53 | 2.68 | -3.07 | -- | 0.0708 | 23.5 | 21.39 | 783 | 2.04 | -- |
| 275.46 | 0.773 | 7.63 | 0.475 | 4.04 | 3.15 | -- | -0.605 | 0.0941 | 23.9 | 20.96 | 725 | 0.785 | -- |
| 275.25 | 0.773 | 7.63 | 0.553 | 4.26 | 3.27 | -- | -- | 0.138 | 0.0973 | 21 | 672 | 0.0305 | -- |
| 707.23 | 1.24 | 7.64 | -0.131 | -- | 2.46 | -- | -0.0531 | -0.119 | -- | 20 | 2928 | 2.09 | -- |
| 456.65 | 1 | 7.46 | 1.31 | 2.39 | -- | -3.42 | -- | 0.743 | 0.137 | -- | -- | -- | -- |
| 507.34 | 1.05 | 7.45 | 1.45 | 2.4 | -- | -- | -- | 0.759 | 0.161 | -- | -- | -- | -- |
| 588.96 | 1.14 | 7.47 | 1.14 | -- | 2.15 | -- | -- | 0.51 | -- | 19.61 | 3260 | 4.88 | -- |
| 283.27 | 0.784 | 7.65 | -- | 4.38 | 3.5 | -- | -- | 0.0333 | 0.161 | 20.96 | 683 | 6.86e-07 | -- |
| 869.58 | 1.38 | 7.4 | 1.52 | -- | -- | -- | -- | 0.794 | -- | -- | -- | -- | -- |
| 562.53 | 1.11 | 7.49 | -- | 2.42 | -- | -- | -- | 0.597 | 0.194 | -- | -- | -- | -- |
| 542.22 | 1.08 | 7.71 | -- | -- | 4.04 | -- | -- | -0.127 | -- | 20 | 3588 | 10.8 | -- |
| 930.89 | 1.42 | 7.44 | -- | -- | -- | -- | -- | 0.625 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance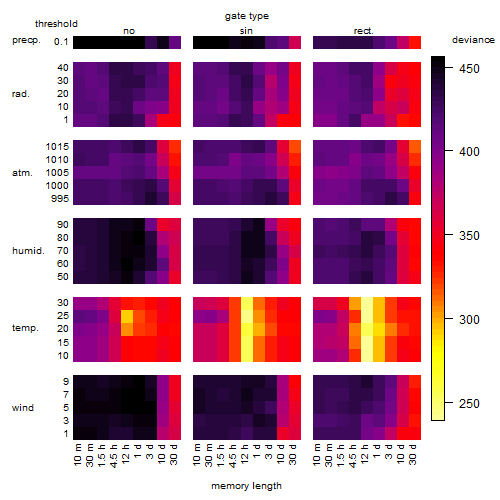
|
Histogram 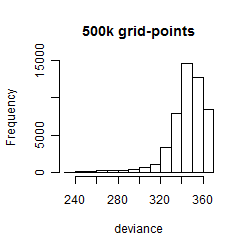
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 238.89 | temperature | 25 | 720 | < th | dose dependent | rect. | 22 | 10 |
| 4 | 239.88 | temperature | 15 | 720 | > th | dose dependent | rect. | 23 | 7 |
| 22 | 244.19 | temperature | 25 | 720 | < th | dose dependent | sin | 8 | NA |
| 35 | 248.13 | temperature | 15 | 720 | > th | dose dependent | rect. | 23 | 10 |
| 68 | 253.34 | temperature | 15 | 720 | > th | dose dependent | sin | 9 | NA |
| 81 | 254.47 | temperature | 15 | 720 | > th | dose dependent | rect. | 21 | 14 |
| 90 | 255.28 | temperature | 20 | 720 | > th | dose independent | rect. | 23 | 8 |
| 93 | 255.58 | temperature | 15 | 720 | > th | dose dependent | rect. | 21 | 12 |
| 97 | 255.81 | temperature | 20 | 720 | < th | dose independent | rect. | 3 | 4 |
| 104 | 256.27 | temperature | 20 | 720 | > th | dose independent | rect. | 23 | 10 |
| 105 | 256.34 | temperature | 20 | 720 | < th | dose independent | rect. | 1 | 6 |
| 106 | 256.45 | temperature | 20 | 720 | < th | dose independent | rect. | 23 | 9 |
| 108 | 256.50 | temperature | 20 | 720 | > th | dose independent | rect. | 1 | 6 |
| 146 | 259.57 | temperature | 20 | 720 | > th | dose independent | sin | 8 | NA |
| 166 | 260.63 | temperature | 20 | 720 | < th | dose independent | sin | 8 | NA |
| 173 | 260.88 | temperature | 25 | 720 | < th | dose dependent | rect. | 3 | 2 |
| 185 | 261.41 | temperature | 20 | 720 | > th | dose independent | rect. | 23 | 12 |
| 230 | 264.15 | temperature | 20 | 720 | > th | dose independent | rect. | 21 | 14 |
| 286 | 266.97 | temperature | 20 | 720 | < th | dose independent | rect. | 5 | 1 |
| 762 | 289.47 | temperature | 20 | 1440 | > th | dose independent | rect. | 5 | 1 |
| 830 | 291.76 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 870 | 293.29 | temperature | 15 | 1440 | > th | dose dependent | rect. | 4 | 1 |