Os06g0633800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Amino acid/polyamine transporter II family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os06g0633800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Amino acid/polyamine transporter II family protein.
|
|
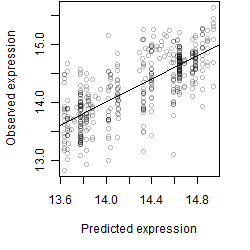
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 49.57 | 0.331 | 14.2 | -0.0151 | 1.23 | 0.583 | -0.0365 | 0.486 | 0.0996 | 9.58 | 24.1 | 1194 | -- | -- |
| 49.68 | 0.328 | 14.2 | -0.131 | 1.24 | 0.628 | -0.0283 | -- | 0.0644 | 9.59 | 24.31 | 1206 | -- | -- |
| 49.57 | 0.328 | 14.2 | -0.0106 | 1.23 | 0.578 | -- | 0.504 | 0.101 | 9.58 | 24.1 | 1200 | -- | -- |
| 49.68 | 0.328 | 14.2 | -0.142 | 1.23 | 0.626 | -- | -- | 0.0696 | 9.58 | 24.27 | 1205 | -- | -- |
| 131.76 | 0.535 | 14.3 | -0.127 | -- | 0.509 | -- | 0.189 | 0.0629 | -- | 22.35 | 5817 | -- | -- |
| 62.51 | 0.371 | 14.2 | 0.0559 | 1.16 | -- | -0.0843 | -- | 0.159 | 9.28 | -- | -- | -- | -- |
| 62.53 | 0.37 | 14.2 | 0.0527 | 1.16 | -- | -- | -- | 0.159 | 9.26 | -- | -- | -- | -- |
| 129.2 | 0.529 | 14.2 | -0.0647 | -- | 0.473 | -- | -- | 0.213 | -- | 29.57 | 5820 | -- | -- |
| 50.18 | 0.33 | 14.2 | -- | 1.24 | 0.606 | -- | -- | 0.0857 | 9.56 | 24.29 | 1198 | -- | -- |
| 140.85 | 0.555 | 14.2 | 0.0478 | -- | -- | -- | -- | 0.164 | -- | -- | -- | -- | -- |
| 62.61 | 0.37 | 14.2 | -- | 1.16 | -- | -- | -- | 0.153 | 9.26 | -- | -- | -- | -- |
| 129.37 | 0.53 | 14.2 | -- | -- | 0.479 | -- | -- | 0.229 | -- | 29.6 | 5810 | -- | -- |
| 140.91 | 0.554 | 14.2 | -- | -- | -- | -- | -- | 0.158 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 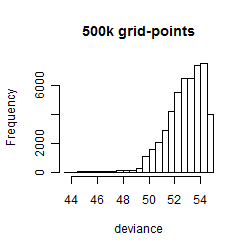
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 43.72 | temperature | 15 | 720 | > th | dose dependent | rect. | 17 | 7 |
| 11 | 44.31 | temperature | 25 | 720 | > th | dose dependent | rect. | 21 | 1 |
| 13 | 44.42 | temperature | 25 | 720 | > th | dose dependent | rect. | 20 | 3 |
| 38 | 44.95 | temperature | 10 | 720 | > th | dose dependent | sin | 15 | NA |
| 96 | 45.69 | temperature | 10 | 720 | > th | dose dependent | rect. | 13 | 16 |
| 98 | 45.72 | temperature | 25 | 720 | > th | dose independent | rect. | 17 | 7 |
| 138 | 46.08 | temperature | 25 | 720 | < th | dose independent | rect. | 18 | 5 |
| 208 | 46.54 | temperature | 15 | 270 | > th | dose dependent | sin | 10 | NA |
| 218 | 46.60 | temperature | 30 | 720 | < th | dose dependent | rect. | 17 | 9 |
| 388 | 47.80 | temperature | 30 | 720 | < th | dose dependent | rect. | 18 | 3 |
| 407 | 47.87 | temperature | 25 | 720 | > th | dose independent | sin | 15 | NA |
| 498 | 48.35 | temperature | 30 | 720 | < th | dose dependent | sin | 15 | NA |
| 506 | 48.40 | temperature | 25 | 43200 | > th | dose dependent | rect. | 22 | 1 |
| 565 | 48.72 | temperature | 30 | 270 | < th | dose dependent | sin | 10 | NA |
| 592 | 48.82 | temperature | 30 | 43200 | < th | dose independent | rect. | 18 | 13 |
| 612 | 48.87 | temperature | 30 | 43200 | < th | dose independent | rect. | 18 | 11 |
| 617 | 48.89 | temperature | 30 | 43200 | < th | dose independent | rect. | 18 | 2 |
| 618 | 48.89 | temperature | 30 | 43200 | < th | dose independent | rect. | 18 | 4 |
| 619 | 48.89 | temperature | 30 | 43200 | < th | dose independent | rect. | 18 | 9 |
| 623 | 48.90 | temperature | 30 | 43200 | < th | dose independent | rect. | 18 | 7 |
| 667 | 49.03 | temperature | 30 | 43200 | > th | dose independent | rect. | 18 | 1 |
| 691 | 49.06 | temperature | 25 | 720 | < th | dose independent | sin | 15 | NA |
| 698 | 49.09 | wind | 5 | 43200 | > th | dose independent | rect. | 20 | 2 |
| 748 | 49.24 | wind | 5 | 43200 | > th | dose independent | rect. | 20 | 5 |
| 757 | 49.26 | wind | 5 | 43200 | < th | dose independent | rect. | 20 | 2 |
| 778 | 49.32 | temperature | 25 | 43200 | > th | dose independent | rect. | 22 | 1 |
| 814 | 49.37 | wind | 5 | 43200 | < th | dose independent | rect. | 20 | 5 |
| 817 | 49.38 | temperature | 25 | 43200 | < th | dose independent | rect. | 22 | 2 |
| 873 | 49.44 | temperature | 20 | 10 | > th | dose dependent | rect. | 21 | 9 |
| 979 | 49.57 | temperature | 30 | 43200 | > th | dose dependent | sin | 13 | NA |
| 981 | 49.57 | temperature | 30 | 43200 | > th | dose dependent | rect. | 14 | 5 |