Os06g0475400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF563 family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os06g0475400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF563 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
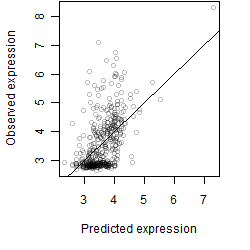
Dependence on each variable

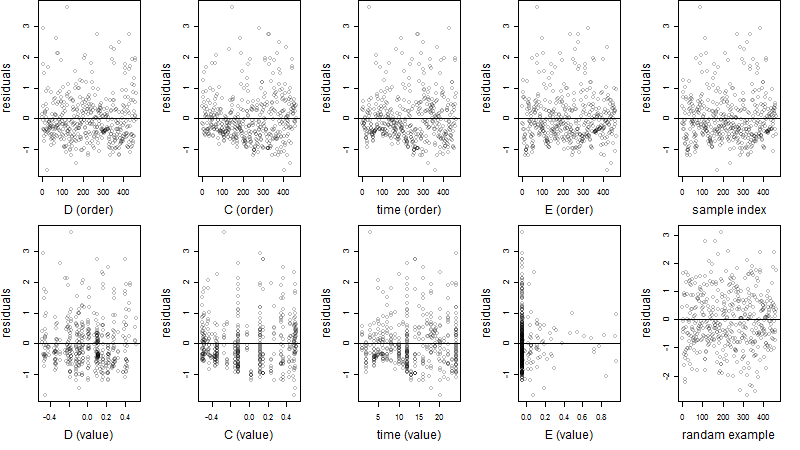
Residual plot
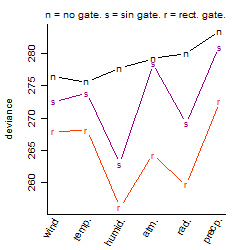
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = < th, dose dependency = dose dependent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 250.11 | 0.743 | 3.56 | 0.264 | 1.07 | 1.37 | 2.63 | -17 | 0.369 | 18.8 | 77.49 | 6 | 16.5 | 17.5 |
| 280.67 | 0.786 | 3.53 | 0.685 | 0.963 | 0.48 | 2.51 | -- | 0.332 | 18 | 177.8 | 2 | 14.4 | 18.6 |
| 264.97 | 0.758 | 3.57 | 0.26 | 0.951 | 1.53 | -- | -16.3 | 0.381 | 18.9 | 76.34 | 4 | 16.6 | 17.4 |
| 294.59 | 0.799 | 3.52 | 0.641 | 1.02 | 0.592 | -- | -- | 0.353 | 17.1 | 287.8 | 57 | 14.3 | 18.9 |
| 317.12 | 0.829 | 3.53 | 0.191 | -- | 2.35 | -- | -17.1 | 0.384 | -- | 76.53 | 3 | 16.6 | 17.4 |
| 288.03 | 0.796 | 3.52 | 0.717 | 1.05 | -- | 2.52 | -- | 0.401 | 18.8 | -- | -- | -- | -- |
| 304.21 | 0.817 | 3.51 | 0.675 | 1.05 | -- | -- | -- | 0.408 | 18.8 | -- | -- | -- | -- |
| 322.91 | 0.837 | 3.48 | 0.786 | -- | 1.32 | -- | -- | 0.386 | -- | 96.18 | 239 | 13 | 18.9 |
| 305.09 | 0.814 | 3.53 | -- | 1.07 | 0.644 | -- | -- | 0.273 | 16.8 | 825.2 | 108 | 13.6 | 18.9 |
| 362.29 | 0.889 | 3.49 | 0.652 | -- | -- | -- | -- | 0.387 | -- | -- | -- | -- | -- |
| 316.2 | 0.832 | 3.53 | -- | 1.05 | -- | -- | -- | 0.333 | 18.9 | -- | -- | -- | -- |
| 314.6 | 0.826 | 3.52 | -- | -- | -0.961 | -- | -- | 0.287 | -- | 870.2 | 655 | 14.7 | 20.9 |
| 373.49 | 0.902 | 3.51 | -- | -- | -- | -- | -- | 0.315 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 256.22 | humidity | 80 | 10 | < th | dose dependent | rect. | 16 | 17 |
| 3 | 257.49 | humidity | 80 | 90 | < th | dose dependent | rect. | 22 | 11 |
| 5 | 257.92 | humidity | 70 | 90 | < th | dose independent | rect. | 6 | 3 |
| 22 | 258.37 | humidity | 80 | 90 | < th | dose dependent | rect. | 16 | 17 |
| 23 | 258.43 | humidity | 80 | 90 | < th | dose dependent | rect. | 5 | 4 |
| 30 | 259.09 | humidity | 70 | 90 | < th | dose independent | rect. | 16 | 17 |
| 32 | 259.67 | radiation | 30 | 90 | > th | dose dependent | rect. | 7 | 1 |
| 49 | 259.80 | radiation | 30 | 90 | > th | dose independent | rect. | 7 | 1 |
| 77 | 260.03 | radiation | 10 | 30 | > th | dose dependent | rect. | 16 | 16 |
| 195 | 261.25 | radiation | 20 | 1440 | > th | dose dependent | rect. | 6 | 1 |
| 282 | 261.48 | humidity | 80 | 1440 | < th | dose dependent | rect. | 6 | 1 |
| 319 | 261.51 | humidity | 70 | 10 | < th | dose independent | rect. | 7 | 1 |
| 394 | 261.51 | humidity | 70 | 10 | < th | dose dependent | rect. | 7 | 1 |
| 506 | 261.56 | humidity | 80 | 1440 | > th | dose independent | rect. | 6 | 1 |
| 524 | 261.71 | humidity | 80 | 1440 | < th | dose independent | rect. | 6 | 1 |
| 541 | 261.78 | humidity | 80 | 1440 | < th | dose dependent | rect. | 24 | 7 |
| 542 | 261.80 | humidity | 70 | 4320 | < th | dose dependent | rect. | 20 | 11 |
| 555 | 261.82 | humidity | 70 | 4320 | < th | dose independent | rect. | 20 | 11 |
| 616 | 261.92 | humidity | 70 | 1440 | > th | dose independent | rect. | 20 | 11 |
| 688 | 262.31 | humidity | 70 | 10 | < th | dose independent | rect. | 16 | 17 |
| 783 | 262.65 | humidity | 80 | 1440 | > th | dose independent | rect. | 3 | 4 |
| 804 | 262.73 | humidity | 70 | 1440 | > th | dose independent | rect. | 23 | 9 |
| 843 | 262.90 | humidity | 80 | 10 | < th | dose dependent | sin | 11 | NA |
| 857 | 263.03 | humidity | 70 | 1440 | > th | dose independent | rect. | 2 | 6 |
| 950 | 264.29 | atmosphere | 1005 | 10 | > th | dose dependent | rect. | 7 | 1 |