Os06g0334100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Non-protein coding transcript, putative npRNA.

FiT-DB / Search/ Help/ Sample detail
|
Os06g0334100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Non-protein coding transcript, putative npRNA.
|
|
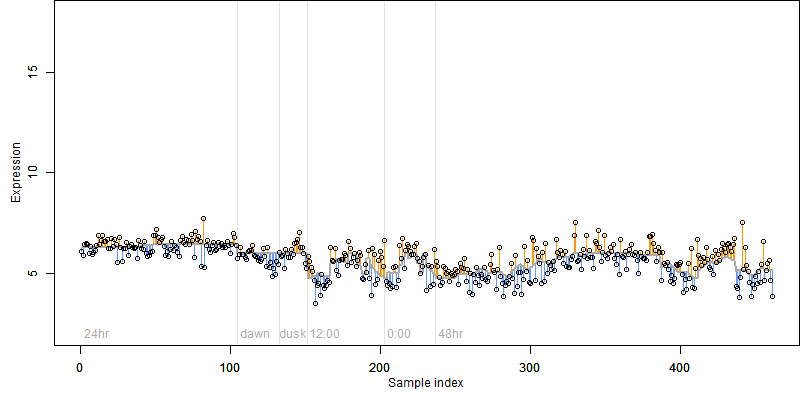
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

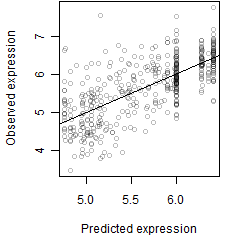 __
__
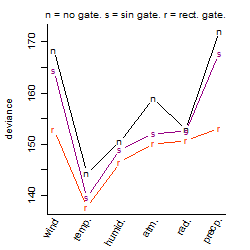
Dependence on each variable

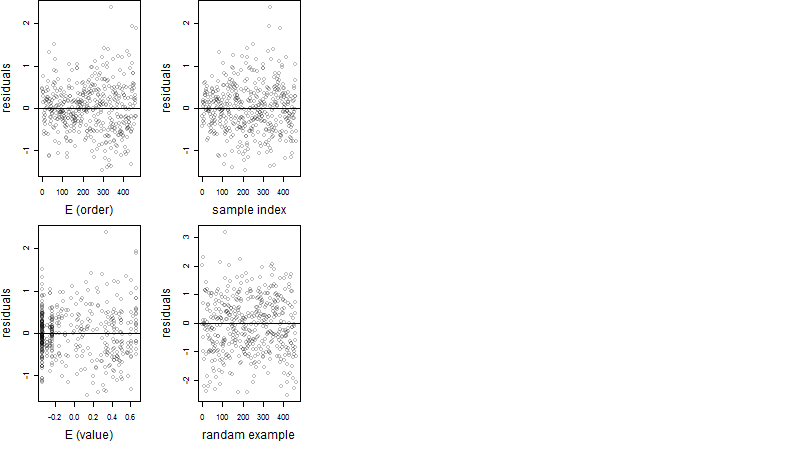
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 142.71 | 0.561 | 5.58 | 0.12 | 0.281 | -1.38 | 0.399 | 0.219 | 0.467 | 19.7 | 24.8 | 1386 | -- | -- |
| 142.82 | 0.557 | 5.57 | 0.155 | 0.266 | -1.36 | 0.377 | -- | 0.477 | 19.7 | 24.77 | 1381 | -- | -- |
| 143.16 | 0.557 | 5.58 | 0.114 | 0.282 | -1.38 | -- | 0.209 | 0.469 | 19.7 | 24.79 | 1381 | -- | -- |
| 143.05 | 0.557 | 5.56 | 0.183 | 0.271 | -1.35 | -- | -- | 0.489 | 19 | 24.8 | 1302 | -- | -- |
| 143.48 | 0.558 | 5.56 | 0.0677 | -- | -1.44 | -- | 0.18 | 0.496 | -- | 24.7 | 2147 | -- | -- |
| 199.8 | 0.663 | 5.46 | 0.651 | 0.264 | -- | 0.327 | -- | 0.969 | 21.8 | -- | -- | -- | -- |
| 200.21 | 0.663 | 5.46 | 0.638 | 0.262 | -- | -- | -- | 0.968 | 22 | -- | -- | -- | -- |
| 143.21 | 0.557 | 5.57 | 0.143 | -- | -1.18 | -- | -- | 0.461 | -- | 23.6 | 2216 | -- | -- |
| 143.88 | 0.559 | 5.57 | -- | 0.278 | -1.41 | -- | -- | 0.453 | 18.8 | 24.8 | 1302 | -- | -- |
| 204.34 | 0.668 | 5.45 | 0.641 | -- | -- | -- | -- | 0.968 | -- | -- | -- | -- | -- |
| 210.92 | 0.679 | 5.48 | -- | 0.265 | -- | -- | -- | 0.897 | 22.2 | -- | -- | -- | -- |
| 143.48 | 0.558 | 5.58 | -- | -- | -1.25 | -- | -- | 0.419 | -- | 23.65 | 2215 | -- | -- |
| 215.17 | 0.685 | 5.47 | -- | -- | -- | -- | -- | 0.897 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance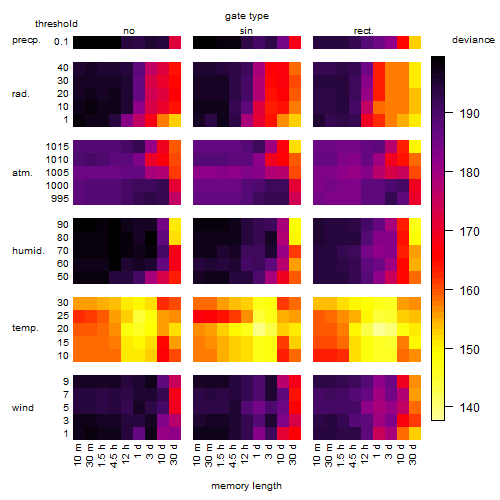
|
Histogram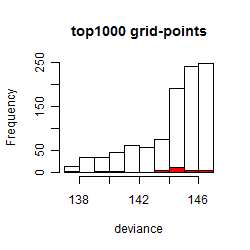 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 137.67 | temperature | 20 | 1440 | > th | dose dependent | rect. | 21 | 10 |
| 63 | 139.55 | temperature | 20 | 1440 | > th | dose dependent | sin | 10 | NA |
| 109 | 140.71 | temperature | 20 | 1440 | > th | dose dependent | rect. | 5 | 1 |
| 279 | 143.68 | temperature | 25 | 1440 | < th | dose independent | rect. | 2 | 22 |
| 287 | 143.74 | temperature | 25 | 1440 | < th | dose independent | rect. | 15 | 22 |
| 293 | 143.82 | temperature | 25 | 1440 | < th | dose independent | rect. | 6 | 21 |
| 333 | 144.07 | temperature | 25 | 1440 | > th | dose independent | rect. | 4 | 21 |
| 342 | 144.11 | temperature | 25 | 1440 | < th | dose independent | sin | 17 | NA |
| 375 | 144.33 | temperature | 25 | 1440 | < th | dose independent | no | NA | NA |
| 396 | 144.44 | temperature | 25 | 1440 | < th | dose independent | rect. | 9 | 23 |
| 426 | 144.61 | temperature | 25 | 4320 | < th | dose dependent | rect. | 21 | 1 |
| 427 | 144.62 | temperature | 25 | 1440 | < th | dose independent | rect. | 15 | 9 |
| 434 | 144.67 | temperature | 25 | 1440 | > th | dose independent | rect. | 15 | 22 |
| 462 | 144.81 | temperature | 25 | 1440 | < th | dose independent | rect. | 15 | 7 |
| 473 | 144.86 | temperature | 25 | 1440 | > th | dose independent | sin | 17 | NA |
| 503 | 144.98 | temperature | 25 | 1440 | < th | dose independent | rect. | 20 | 23 |
| 516 | 145.04 | temperature | 25 | 1440 | > th | dose independent | no | NA | NA |
| 519 | 145.06 | temperature | 25 | 1440 | > th | dose independent | rect. | 10 | 23 |
| 615 | 145.47 | temperature | 25 | 1440 | > th | dose independent | rect. | 20 | 23 |
| 630 | 145.56 | temperature | 25 | 1440 | > th | dose independent | rect. | 15 | 9 |
| 844 | 146.29 | temperature | 20 | 1440 | > th | dose dependent | no | NA | NA |
| 871 | 146.37 | humidity | 90 | 43200 | > th | dose dependent | rect. | 23 | 1 |
| 896 | 146.42 | temperature | 30 | 1440 | < th | dose dependent | rect. | 18 | 11 |