Os06g0317600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Similarity.
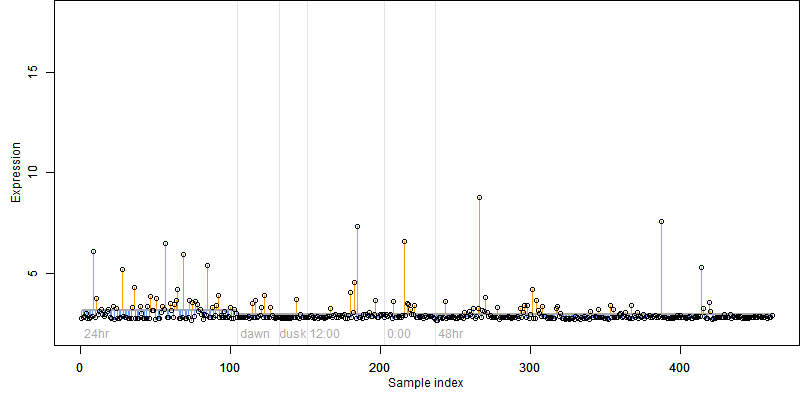

FiT-DB / Search/ Help/ Sample detail
|
Os06g0317600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Similarity.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
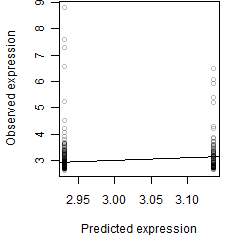
Dependence on each variable

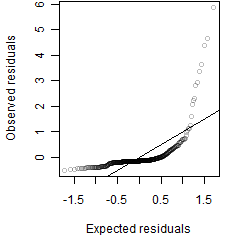
Residual plot

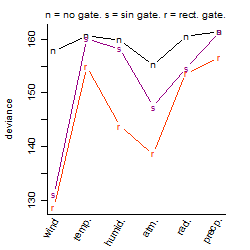
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose dependent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 130.82 | 0.537 | 2.78 | -0.541 | 0.0396 | -20 | -0.717 | -92.5 | 0.234 | 0.686 | 8.687 | 89 | 6.44 | -- |
| 161.16 | 0.596 | 2.93 | -0.0508 | 0.0694 | 0.66 | -0.818 | -- | 0.209 | 21.5 | 8.541 | 418 | 11.2 | -- |
| 130.78 | 0.537 | 2.68 | -1.02 | 0.0505 | -94 | -- | -435 | 0.231 | 22 | 8.687 | 89 | 6.44 | -- |
| 144.51 | 0.564 | 2.92 | 0.0398 | 0.0463 | 3.64 | -- | -- | 0.237 | 0.1 | 7.441 | 2 | 8.4 | -- |
| 130.93 | 0.536 | 2.68 | -1.02 | -- | -94.1 | -- | -435 | 0.231 | -- | 8.687 | 89 | 6.44 | -- |
| 162.04 | 0.597 | 2.94 | -0.0799 | 0.0814 | -- | -0.85 | -- | 0.199 | 21.1 | -- | -- | -- | -- |
| 164.52 | 0.601 | 2.93 | -0.0485 | 0.0942 | -- | -- | -- | 0.201 | 19.7 | -- | -- | -- | -- |
| 132.75 | 0.539 | 2.92 | 0.0615 | -- | 5.58 | -- | -- | 0.235 | -- | 8 | 2 | 9.24 | -- |
| 132.68 | 0.539 | 2.93 | -- | 0.0555 | 5.54 | -- | -- | 0.228 | 22.7 | 8 | 2 | 9.27 | -- |
| 165 | 0.6 | 2.93 | -0.0497 | -- | -- | -- | -- | 0.199 | -- | -- | -- | -- | -- |
| 164.58 | 0.6 | 2.93 | -- | 0.0945 | -- | -- | -- | 0.206 | 19.7 | -- | -- | -- | -- |
| 132.85 | 0.539 | 2.93 | -- | -- | 5.55 | -- | -- | 0.228 | -- | 8 | 2 | 9.24 | -- |
| 165.06 | 0.6 | 2.93 | -- | -- | -- | -- | -- | 0.205 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance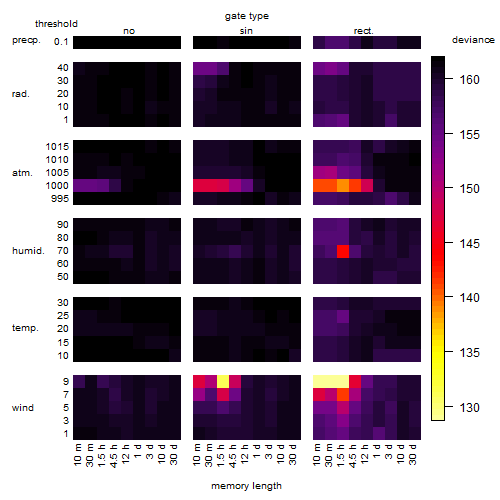
|
Histogram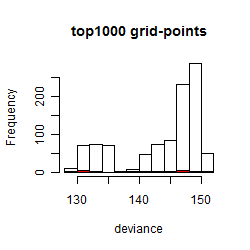 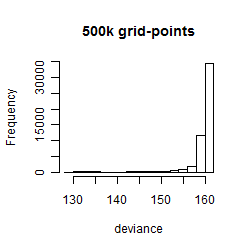
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 128.66 | wind | 9 | 10 | > th | dose independent | rect. | 18 | 1 |
| 4 | 128.66 | wind | 9 | 90 | > th | dose dependent | rect. | 17 | 1 |
| 5 | 128.66 | wind | 9 | 10 | > th | dose dependent | rect. | 18 | 1 |
| 10 | 130.27 | wind | 9 | 90 | > th | dose dependent | rect. | 16 | 18 |
| 27 | 130.75 | wind | 9 | 90 | > th | dose independent | rect. | 16 | 6 |
| 59 | 131.03 | wind | 9 | 90 | < th | dose independent | sin | 12 | NA |
| 60 | 131.12 | wind | 9 | 90 | > th | dose independent | sin | 12 | NA |
| 81 | 132.29 | wind | 9 | 90 | > th | dose dependent | sin | 13 | NA |
| 225 | 138.66 | atmosphere | 1000 | 90 | < th | dose independent | rect. | 8 | 3 |
| 237 | 140.87 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 9 | 1 |
| 252 | 141.24 | atmosphere | 1000 | 270 | < th | dose independent | rect. | 6 | 1 |
| 276 | 141.98 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 2 | 9 |
| 287 | 142.20 | atmosphere | 1000 | 90 | < th | dose dependent | rect. | 9 | 1 |
| 343 | 143.58 | atmosphere | 1000 | 270 | < th | dose dependent | rect. | 6 | 1 |
| 347 | 143.78 | humidity | 70 | 90 | < th | dose independent | rect. | 16 | 1 |
| 393 | 145.68 | wind | 9 | 10 | > th | dose independent | rect. | 17 | 5 |
| 555 | 146.97 | wind | 9 | 10 | > th | dose independent | sin | 12 | NA |
| 577 | 147.22 | wind | 9 | 10 | < th | dose independent | sin | 12 | NA |
| 598 | 147.38 | atmosphere | 1000 | 30 | < th | dose independent | sin | 5 | NA |
| 599 | 147.39 | atmosphere | 1000 | 30 | > th | dose independent | sin | 5 | NA |
| 681 | 148.28 | atmosphere | 1000 | 720 | < th | dose dependent | rect. | 22 | 4 |
| 850 | 149.69 | wind | 7 | 10 | > th | dose dependent | rect. | 17 | 5 |
| 965 | 150.11 | atmosphere | 1000 | 720 | < th | dose independent | rect. | 22 | 4 |