Os06g0299100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Glucose/ribitol dehydrogenase family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os06g0299100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Glucose/ribitol dehydrogenase family protein.
|
|
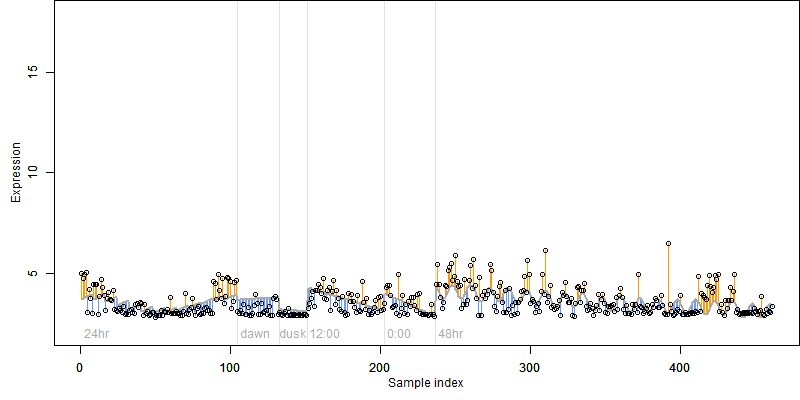
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
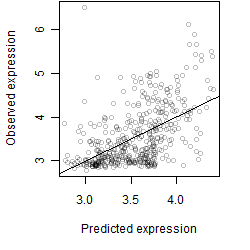
Dependence on each variable

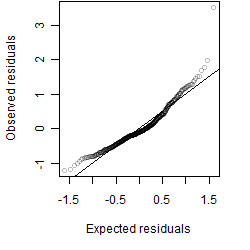
Residual plot
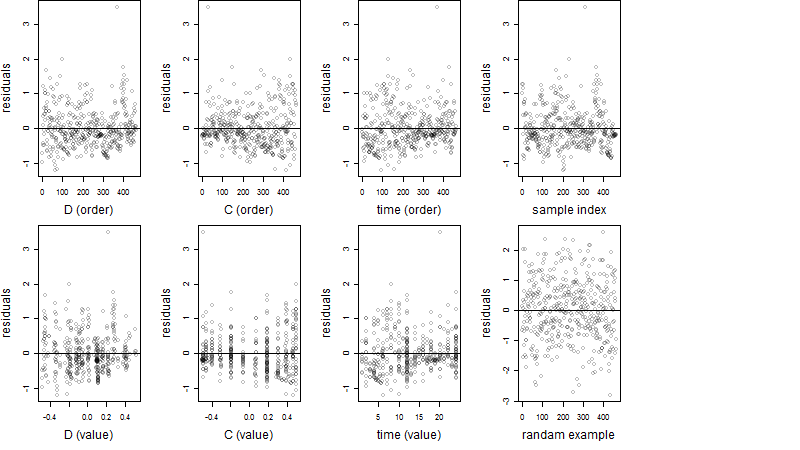
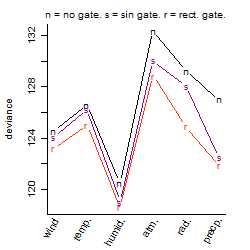
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 120.39 | 0.516 | 3.56 | -1.2 | 0.732 | 0.771 | -1.26 | 1.39 | -0.177 | 7.11 | 60.19 | 1396 | -- | -- |
| 122.72 | 0.516 | 3.56 | -1.02 | 0.695 | 0.796 | -1.2 | -- | -0.172 | 7.5 | 48.99 | 1530 | -- | -- |
| 124.54 | 0.52 | 3.56 | -1.22 | 0.731 | 0.751 | -- | 1.37 | -0.179 | 7.23 | 58.64 | 1410 | -- | -- |
| 126.65 | 0.524 | 3.55 | -1.04 | 0.68 | 0.823 | -- | -- | -0.164 | 7.82 | 49 | 1572 | -- | -- |
| 133.87 | 0.539 | 3.56 | -1.1 | -- | 1.11 | -- | 0.151 | -0.106 | -- | 70.78 | 649 | -- | -- |
| 139.25 | 0.553 | 3.57 | -1.06 | 0.706 | -- | -1.24 | -- | -0.206 | 7.28 | -- | -- | -- | -- |
| 143.33 | 0.561 | 3.56 | -1.08 | 0.703 | -- | -- | -- | -0.203 | 7.5 | -- | -- | -- | -- |
| 133.75 | 0.539 | 3.56 | -1.1 | -- | 1.04 | -- | -- | -0.11 | -- | 71.99 | 620 | -- | -- |
| 155.28 | 0.58 | 3.53 | -- | 0.688 | 0.837 | -- | -- | -0.0516 | 7.7 | 41.56 | 1504 | -- | -- |
| 169.72 | 0.609 | 3.58 | -1.07 | -- | -- | -- | -- | -0.192 | -- | -- | -- | -- | -- |
| 174.27 | 0.618 | 3.54 | -- | 0.693 | -- | -- | -- | -0.0822 | 7.6 | -- | -- | -- | -- |
| 164.55 | 0.597 | 3.54 | -- | -- | 1.1 | -- | -- | 0.000465 | -- | 67 | 685 | -- | -- |
| 200.08 | 0.66 | 3.55 | -- | -- | -- | -- | -- | -0.0728 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance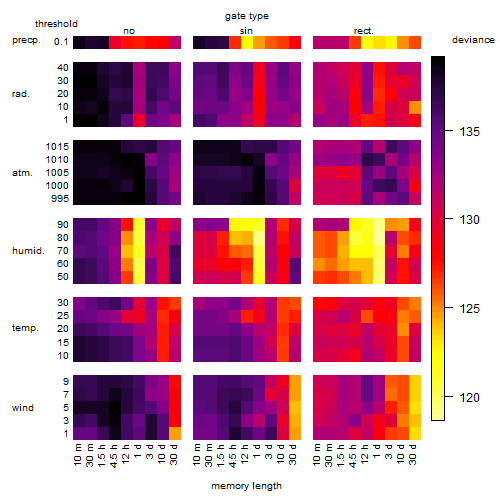
|
Histogram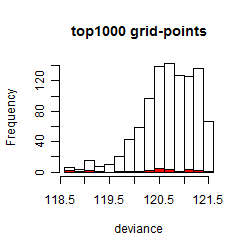 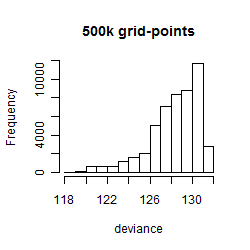
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 118.62 | humidity | 80 | 1440 | < th | dose independent | rect. | 5 | 9 |
| 5 | 118.78 | humidity | 80 | 1440 | > th | dose independent | rect. | 5 | 9 |
| 9 | 119.00 | humidity | 80 | 1440 | < th | dose independent | sin | 3 | NA |
| 11 | 119.03 | humidity | 80 | 1440 | > th | dose independent | sin | 3 | NA |
| 21 | 119.12 | humidity | 90 | 1440 | < th | dose dependent | rect. | 17 | 19 |
| 30 | 119.30 | humidity | 50 | 1440 | > th | dose dependent | rect. | 16 | 20 |
| 46 | 119.64 | humidity | 80 | 1440 | > th | dose independent | rect. | 5 | 17 |
| 91 | 119.95 | humidity | 80 | 1440 | < th | dose independent | rect. | 5 | 17 |
| 123 | 120.06 | humidity | 70 | 1440 | > th | dose dependent | rect. | 6 | 8 |
| 173 | 120.22 | humidity | 70 | 1440 | > th | dose dependent | rect. | 5 | 18 |
| 180 | 120.24 | humidity | 60 | 1440 | > th | dose dependent | rect. | 2 | 21 |
| 268 | 120.41 | humidity | 80 | 1440 | > th | dose independent | rect. | 16 | 22 |
| 319 | 120.50 | humidity | 70 | 1440 | > th | dose dependent | sin | 2 | NA |
| 328 | 120.51 | humidity | 60 | 1440 | > th | dose dependent | no | NA | NA |
| 339 | 120.53 | humidity | 80 | 1440 | < th | dose independent | rect. | 16 | 22 |
| 349 | 120.54 | humidity | 60 | 1440 | > th | dose dependent | rect. | 21 | 23 |
| 442 | 120.66 | humidity | 80 | 1440 | > th | dose independent | rect. | 6 | 4 |
| 494 | 120.73 | humidity | 80 | 270 | > th | dose dependent | rect. | 5 | 17 |
| 534 | 120.79 | humidity | 80 | 1440 | > th | dose independent | no | NA | NA |
| 623 | 120.92 | humidity | 80 | 1440 | < th | dose independent | no | NA | NA |
| 759 | 121.14 | humidity | 90 | 1440 | < th | dose dependent | no | NA | NA |
| 780 | 121.18 | humidity | 90 | 1440 | < th | dose dependent | rect. | 2 | 23 |
| 787 | 121.19 | humidity | 90 | 1440 | < th | dose dependent | rect. | 5 | 23 |
| 814 | 121.24 | humidity | 90 | 1440 | < th | dose dependent | rect. | 20 | 23 |
| 901 | 121.37 | humidity | 80 | 1440 | < th | dose dependent | sin | 10 | NA |
| 935 | 121.41 | humidity | 90 | 1440 | < th | dose dependent | sin | 7 | NA |