Os06g0260500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to FAE1.


FiT-DB / Search/ Help/ Sample detail
|
Os06g0260500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to FAE1.
|
|
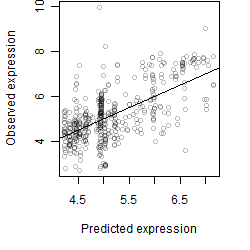
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
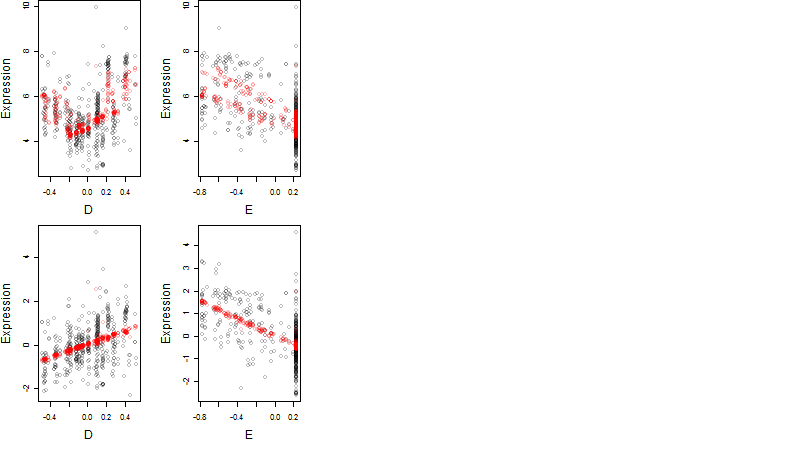
Dependence on each variable
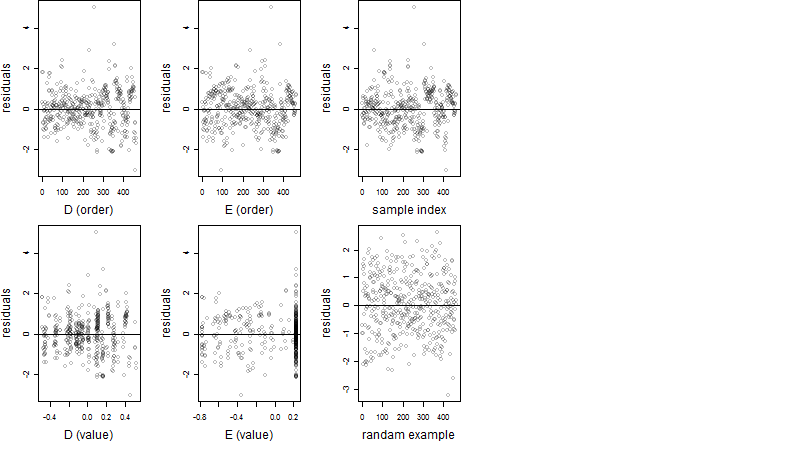
Residual plot
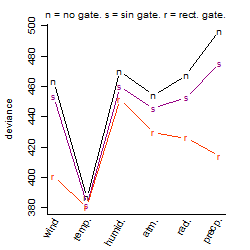
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 375.73 | 0.911 | 5.3 | 1.3 | 0.143 | -2.46 | 0.935 | -2.35 | -0.369 | 8.49 | 21.5 | 2085 | -- | -- |
| 380.53 | 0.909 | 5.23 | 1.6 | 0.317 | -2.1 | 1.41 | -- | -0.25 | 8.3 | 22.2 | 1455 | -- | -- |
| 378.33 | 0.906 | 5.31 | 1.29 | 0.157 | -2.47 | -- | -2.46 | -0.375 | 7.26 | 21.51 | 2084 | -- | -- |
| 386.32 | 0.915 | 5.22 | 1.67 | 0.327 | -2.14 | -- | -- | -0.236 | 7.41 | 22.2 | 1429 | -- | -- |
| 379.56 | 0.907 | 5.31 | 1.3 | -- | -2.5 | -- | -2.5 | -0.367 | -- | 21.51 | 2084 | -- | -- |
| 544.89 | 1.09 | 5.37 | 0.853 | 0.303 | -- | 1.3 | -- | -0.926 | 8.53 | -- | -- | -- | -- |
| 550.34 | 1.1 | 5.37 | 0.895 | 0.299 | -- | -- | -- | -0.926 | 8.72 | -- | -- | -- | -- |
| 388.23 | 0.918 | 5.21 | 1.55 | -- | -1.92 | -- | -- | -0.259 | -- | 22.2 | 1022 | -- | -- |
| 445.86 | 0.983 | 5.28 | -- | 0.161 | -1.61 | -- | -- | -0.517 | 6.97 | 22.3 | 978 | -- | -- |
| 555.39 | 1.1 | 5.38 | 0.895 | -- | -- | -- | -- | -0.923 | -- | -- | -- | -- | -- |
| 571.44 | 1.12 | 5.39 | -- | 0.302 | -- | -- | -- | -1.03 | 8.49 | -- | -- | -- | -- |
| 445.98 | 0.984 | 5.29 | -- | -- | -1.6 | -- | -- | -0.53 | -- | 22.35 | 797 | -- | -- |
| 576.54 | 1.12 | 5.4 | -- | -- | -- | -- | -- | -1.02 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 380.48 | temperature | 25 | 1440 | < th | dose dependent | rect. | 13 | 16 |
| 16 | 381.56 | temperature | 20 | 1440 | > th | dose dependent | rect. | 1 | 2 |
| 23 | 381.73 | temperature | 25 | 1440 | < th | dose dependent | sin | 15 | NA |
| 86 | 384.98 | temperature | 25 | 4320 | < th | dose dependent | rect. | 1 | 1 |
| 96 | 385.29 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 1 |
| 101 | 385.47 | temperature | 25 | 720 | < th | dose dependent | rect. | 24 | 23 |
| 104 | 385.65 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 6 |
| 126 | 386.39 | temperature | 25 | 720 | < th | dose dependent | rect. | 13 | 23 |
| 127 | 386.46 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 4 |
| 131 | 386.52 | temperature | 25 | 720 | < th | dose dependent | rect. | 16 | 23 |
| 148 | 386.78 | temperature | 10 | 4320 | > th | dose dependent | rect. | 1 | 1 |
| 150 | 386.79 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 183 | 387.47 | temperature | 25 | 720 | < th | dose dependent | rect. | 6 | 23 |
| 202 | 387.71 | temperature | 25 | 720 | < th | dose dependent | rect. | 19 | 23 |
| 266 | 388.59 | temperature | 25 | 4320 | < th | dose dependent | rect. | 3 | 1 |
| 324 | 389.43 | temperature | 20 | 1440 | < th | dose independent | rect. | 5 | 1 |
| 418 | 390.55 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 14 |
| 458 | 391.09 | temperature | 20 | 1440 | > th | dose independent | rect. | 5 | 1 |
| 570 | 392.23 | temperature | 20 | 4320 | > th | dose independent | rect. | 22 | 1 |