Os06g0224700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Mitochodrial transcription termination factor-related family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os06g0224700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Mitochodrial transcription termination factor-related family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

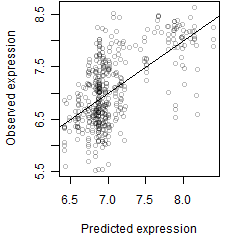 __
__

Dependence on each variable

Residual plot
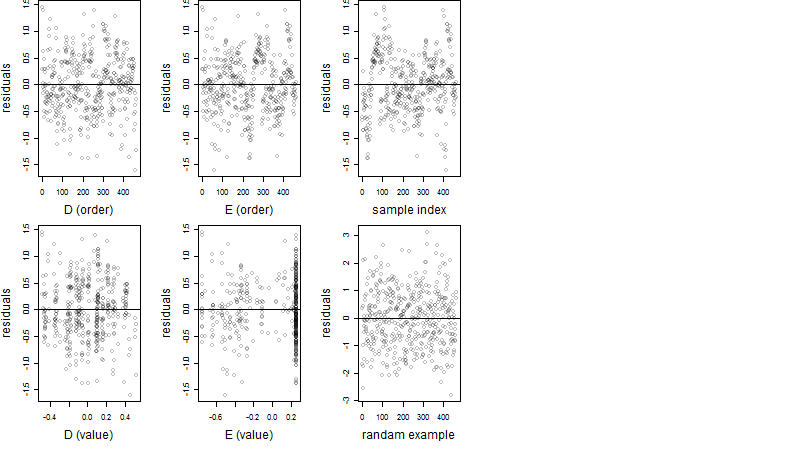
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 112.04 | 0.497 | 6.99 | 1.58 | 0.513 | -1.11 | -0.181 | 0.4 | 0.282 | 1.89 | 22.3 | 990 | -- | -- |
| 112.47 | 0.494 | 7 | 1.45 | 0.513 | -1.11 | -0.095 | -- | 0.25 | 1.93 | 22.28 | 992 | -- | -- |
| 112.15 | 0.493 | 6.99 | 1.56 | 0.513 | -1.11 | -- | 0.344 | 0.279 | 1.96 | 22.3 | 992 | -- | -- |
| 112.5 | 0.494 | 7 | 1.45 | 0.514 | -1.11 | -- | -- | 0.251 | 1.95 | 22.28 | 984 | -- | -- |
| 126.58 | 0.524 | 6.98 | 1.68 | -- | -1.06 | -- | 0.621 | 0.313 | -- | 22.4 | 1462 | -- | -- |
| 164.07 | 0.6 | 7.08 | 1.08 | 0.378 | -- | 0.0663 | -- | -0.123 | 3.22 | -- | -- | -- | -- |
| 164.08 | 0.6 | 7.08 | 1.08 | 0.378 | -- | -- | -- | -0.124 | 3.17 | -- | -- | -- | -- |
| 127.69 | 0.526 | 6.99 | 1.46 | -- | -1.05 | -- | -- | 0.276 | -- | 22.7 | 1461 | -- | -- |
| 163.28 | 0.595 | 7.05 | -- | 0.489 | -0.827 | -- | -- | 0.0128 | 2.21 | 22.35 | 943 | -- | -- |
| 172.19 | 0.613 | 7.08 | 1.09 | -- | -- | -- | -- | -0.114 | -- | -- | -- | -- | -- |
| 194.59 | 0.653 | 7.11 | -- | 0.399 | -- | -- | -- | -0.244 | 3.17 | -- | -- | -- | -- |
| 177.46 | 0.62 | 7.06 | -- | -- | -0.713 | -- | -- | -0.0158 | -- | 22.35 | 757 | -- | -- |
| 203.64 | 0.666 | 7.11 | -- | -- | -- | -- | -- | -0.235 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 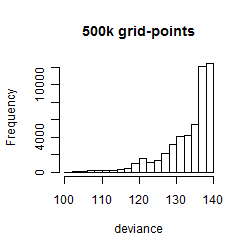
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 101.12 | temperature | 10 | 720 | > th | dose dependent | rect. | 21 | 16 |
| 2 | 101.27 | temperature | 10 | 720 | > th | dose dependent | sin | 6 | NA |
| 7 | 101.83 | temperature | 10 | 720 | > th | dose dependent | rect. | 23 | 16 |
| 23 | 102.79 | temperature | 30 | 720 | < th | dose dependent | rect. | 24 | 14 |
| 25 | 102.91 | temperature | 30 | 720 | < th | dose dependent | sin | 6 | NA |
| 30 | 103.21 | temperature | 10 | 720 | > th | dose dependent | rect. | 1 | 8 |
| 36 | 103.46 | temperature | 10 | 720 | > th | dose dependent | rect. | 1 | 14 |
| 46 | 103.99 | temperature | 10 | 270 | > th | dose dependent | sin | 3 | NA |
| 121 | 106.11 | temperature | 15 | 720 | > th | dose dependent | rect. | 23 | 10 |
| 166 | 106.73 | temperature | 30 | 270 | < th | dose dependent | sin | 2 | NA |
| 175 | 106.93 | temperature | 30 | 270 | < th | dose dependent | rect. | 23 | 22 |
| 428 | 110.30 | temperature | 25 | 720 | < th | dose dependent | rect. | 3 | 4 |
| 541 | 111.55 | temperature | 15 | 270 | > th | dose dependent | rect. | 1 | 23 |
| 631 | 112.53 | temperature | 15 | 270 | > th | dose dependent | rect. | 3 | 23 |
| 659 | 112.78 | temperature | 20 | 270 | > th | dose dependent | rect. | 1 | 21 |
| 708 | 113.17 | temperature | 20 | 10 | > th | dose dependent | rect. | 1 | 23 |
| 767 | 113.73 | temperature | 20 | 10 | > th | dose dependent | rect. | 1 | 19 |
| 771 | 113.76 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 23 |
| 787 | 113.93 | temperature | 20 | 10 | > th | dose dependent | rect. | 3 | 17 |
| 792 | 113.95 | temperature | 30 | 270 | < th | dose dependent | rect. | 5 | 14 |
| 799 | 114.00 | temperature | 20 | 10 | > th | dose dependent | rect. | 1 | 21 |
| 802 | 114.03 | temperature | 15 | 270 | > th | dose dependent | no | NA | NA |
| 804 | 114.03 | temperature | 20 | 10 | > th | dose dependent | rect. | 23 | 23 |
| 817 | 114.14 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 881 | 114.55 | temperature | 30 | 270 | < th | dose dependent | rect. | 7 | 12 |
| 914 | 114.76 | temperature | 30 | 1440 | < th | dose dependent | rect. | 17 | 1 |
| 946 | 114.99 | wind | 9 | 43200 | > th | dose dependent | rect. | 18 | 12 |
| 983 | 115.26 | temperature | 20 | 10 | > th | dose dependent | rect. | 3 | 19 |