Os06g0201200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.

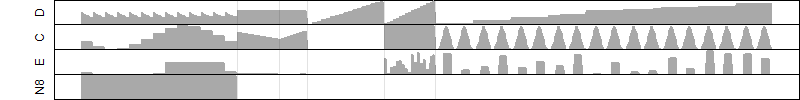
FiT-DB / Search/ Help/ Sample detail
|
Os06g0201200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
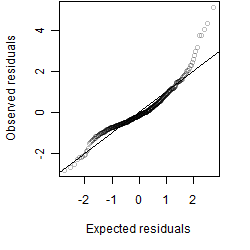
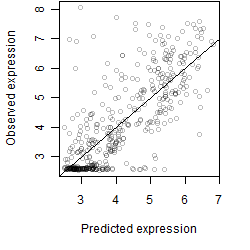 __
__
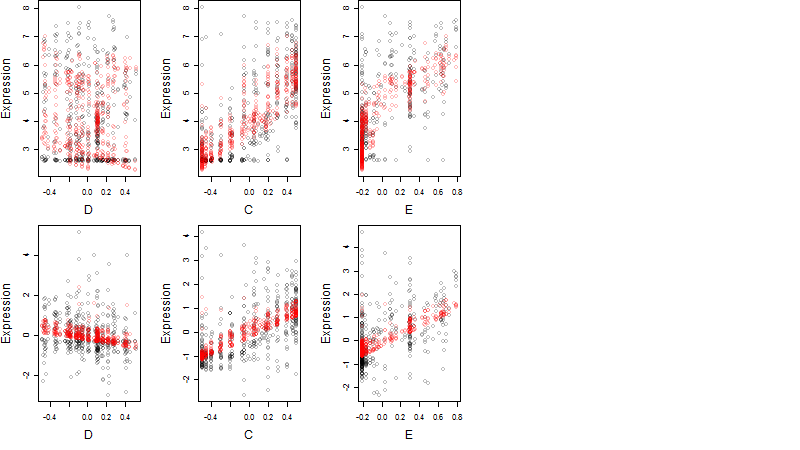
Dependence on each variable
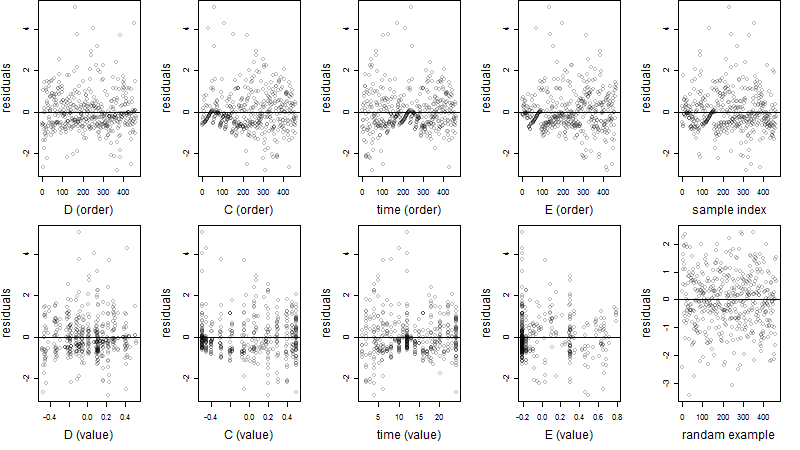
Residual plot
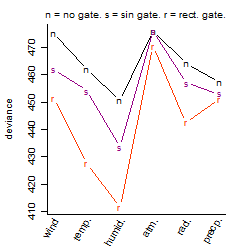
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose dependent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 406.17 | 0.947 | 4.21 | -0.988 | 1.86 | 1.87 | -2.93 | 2.23 | -0.355 | 23.8 | 68.01 | 665 | 19.1 | 0.933 |
| 413.39 | 0.947 | 4.23 | -0.808 | 1.84 | 1.97 | -1.6 | -- | -0.361 | 23.6 | 67.74 | 675 | 19.1 | 0.886 |
| 425.75 | 0.961 | 4.18 | -0.807 | 1.92 | 1.89 | -- | 0.347 | -0.299 | 23.5 | 68 | 669 | 19 | 1.08 |
| 423.35 | 0.958 | 4.22 | -0.763 | 1.82 | 1.97 | -- | -- | -0.364 | 23.6 | 67.13 | 671 | 19 | 1.05 |
| 458.15 | 0.997 | 4.26 | -0.518 | -- | 3.36 | -- | -1.76 | -0.596 | -- | 51.79 | 694 | 17.4 | 3.05 |
| 498.36 | 1.05 | 4.24 | -0.688 | 2.9 | -- | -1.17 | -- | -0.357 | 0.0584 | -- | -- | -- | -- |
| 504.25 | 1.05 | 4.23 | -0.641 | 2.9 | -- | -- | -- | -0.351 | 0.047 | -- | -- | -- | -- |
| 449.38 | 0.987 | 4.19 | -0.647 | -- | 3.59 | -- | -- | -0.422 | -- | 50.1 | 693 | 17.3 | 3.05 |
| 437.99 | 0.975 | 4.22 | -- | 1.8 | 1.99 | -- | -- | -0.304 | 23.5 | 66.92 | 672 | 19 | 1.1 |
| 1033.94 | 1.5 | 4.17 | -0.55 | -- | -- | -- | -- | -0.31 | -- | -- | -- | -- | -- |
| 515.08 | 1.06 | 4.22 | -- | 2.89 | -- | -- | -- | -0.28 | 0.0345 | -- | -- | -- | -- |
| 451.89 | 0.99 | 4.14 | -- | -- | 3.36 | -- | -- | -0.292 | -- | 39.38 | 729 | 16.8 | 2.97 |
| 1041.92 | 1.51 | 4.16 | -- | -- | -- | -- | -- | -0.249 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance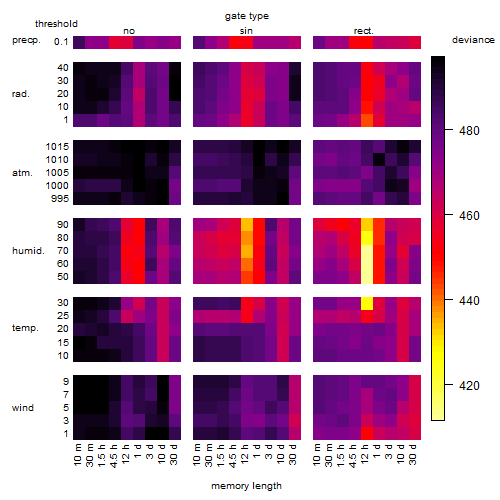
|
Histogram 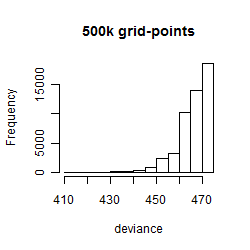
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 411.59 | humidity | 70 | 720 | > th | dose dependent | rect. | 19 | 1 |
| 2 | 411.68 | humidity | 60 | 720 | > th | dose dependent | rect. | 17 | 3 |
| 27 | 426.11 | humidity | 70 | 720 | > th | dose independent | rect. | 16 | 4 |
| 35 | 427.51 | temperature | 30 | 720 | < th | dose independent | rect. | 17 | 3 |
| 45 | 429.50 | temperature | 30 | 720 | < th | dose independent | rect. | 17 | 12 |
| 62 | 431.57 | humidity | 70 | 720 | > th | dose independent | rect. | 16 | 13 |
| 63 | 431.69 | humidity | 90 | 720 | < th | dose dependent | rect. | 15 | 10 |
| 74 | 432.58 | humidity | 80 | 720 | < th | dose independent | rect. | 14 | 8 |
| 76 | 432.69 | humidity | 80 | 720 | > th | dose independent | rect. | 11 | 11 |
| 93 | 433.60 | humidity | 90 | 720 | < th | dose dependent | sin | 14 | NA |
| 96 | 433.67 | humidity | 70 | 720 | > th | dose dependent | rect. | 10 | 12 |
| 122 | 434.96 | humidity | 90 | 720 | < th | dose dependent | rect. | 15 | 17 |
| 126 | 435.01 | humidity | 50 | 720 | > th | dose dependent | rect. | 17 | 11 |
| 130 | 435.13 | humidity | 70 | 720 | > th | dose independent | rect. | 18 | 10 |
| 134 | 435.24 | humidity | 70 | 720 | > th | dose dependent | sin | 19 | NA |
| 207 | 437.77 | humidity | 80 | 720 | > th | dose independent | sin | 19 | NA |
| 258 | 439.07 | humidity | 70 | 720 | < th | dose independent | rect. | 14 | 18 |
| 298 | 439.79 | humidity | 70 | 720 | < th | dose independent | sin | 13 | NA |
| 307 | 439.98 | humidity | 80 | 720 | < th | dose independent | sin | 18 | NA |
| 371 | 441.45 | humidity | 70 | 720 | > th | dose independent | sin | 13 | NA |
| 445 | 442.49 | radiation | 1 | 720 | < th | dose dependent | rect. | 12 | 7 |
| 449 | 442.50 | radiation | 1 | 720 | < th | dose dependent | rect. | 9 | 10 |
| 589 | 444.44 | radiation | 1 | 720 | < th | dose independent | rect. | 17 | 2 |
| 635 | 445.10 | humidity | 90 | 720 | < th | dose independent | rect. | 20 | 4 |
| 649 | 445.25 | humidity | 80 | 720 | < th | dose dependent | rect. | 19 | 2 |
| 686 | 445.62 | humidity | 80 | 720 | > th | dose independent | rect. | 3 | 19 |
| 812 | 446.62 | humidity | 80 | 1440 | < th | dose independent | rect. | 6 | 16 |