Os06g0178700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
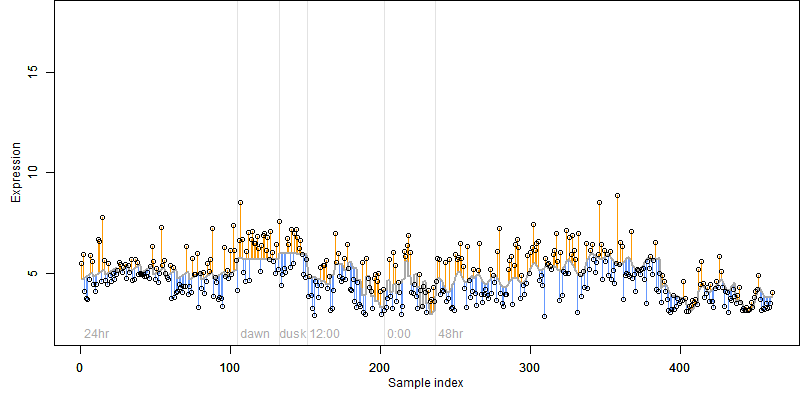
Description : Similar to Iron/ascorbate-dependent oxidoreductase.
FiT-DB / Search/ Help/ Sample detail
|
Os06g0178700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Iron/ascorbate-dependent oxidoreductase.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable
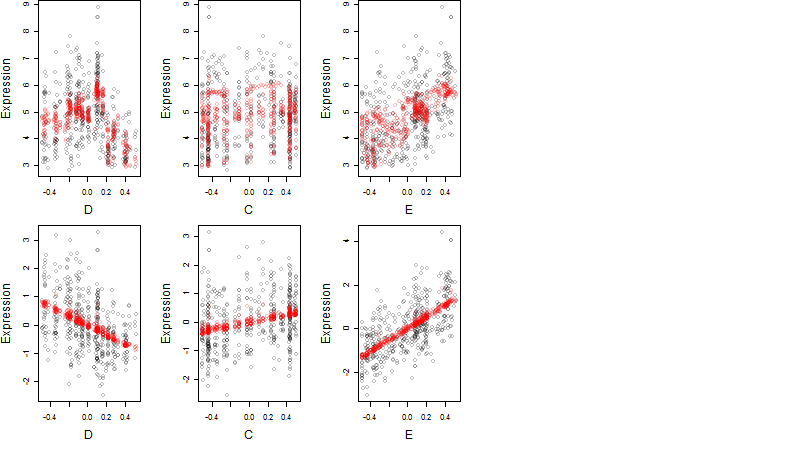
Residual plot
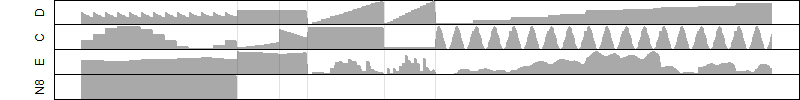
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
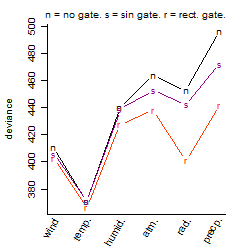
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 363.47 | 0.896 | 5.14 | -3.49 | 0.673 | 3.13 | -0.746 | -7.52 | -0.761 | 14.7 | 24.28 | 1284 | -- | -- |
| 380.27 | 0.908 | 4.94 | -1.58 | 0.647 | 2.56 | -0.434 | -- | -0.441 | 14.8 | 21.11 | 1262 | -- | -- |
| 365.08 | 0.89 | 5.13 | -3.47 | 0.681 | 3.14 | -- | -7.34 | -0.749 | 14.2 | 24.28 | 1274 | -- | -- |
| 380.24 | 0.908 | 4.94 | -1.6 | 0.689 | 2.64 | -- | -- | -0.449 | 13.9 | 20.98 | 1197 | -- | -- |
| 383.27 | 0.912 | 5.13 | -3.54 | -- | 2.81 | -- | -7.17 | -0.713 | -- | 24.75 | 1460 | -- | -- |
| 584.41 | 1.13 | 4.81 | -0.716 | 0.254 | -- | -1.53 | -- | 0.136 | 17.1 | -- | -- | -- | -- |
| 589.34 | 1.14 | 4.81 | -0.715 | 0.27 | -- | -- | -- | 0.13 | 15.2 | -- | -- | -- | -- |
| 396.22 | 0.927 | 4.95 | -1.64 | -- | 2.49 | -- | -- | -0.462 | -- | 20.79 | 1602 | -- | -- |
| 439.15 | 0.976 | 4.88 | -- | 0.662 | 2.17 | -- | -- | -0.165 | 14 | 22.17 | 1197 | -- | -- |
| 593.48 | 1.14 | 4.81 | -0.726 | -- | -- | -- | -- | 0.123 | -- | -- | -- | -- | -- |
| 602.8 | 1.15 | 4.8 | -- | 0.284 | -- | -- | -- | 0.209 | 15.2 | -- | -- | -- | -- |
| 457.79 | 0.997 | 4.88 | -- | -- | 1.93 | -- | -- | -0.172 | -- | 21.9 | 1501 | -- | -- |
| 607.39 | 1.15 | 4.8 | -- | -- | -- | -- | -- | 0.203 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 366.52 | temperature | 20 | 720 | > th | dose dependent | rect. | 16 | 17 |
| 4 | 369.64 | temperature | 25 | 1440 | > th | dose dependent | rect. | 5 | 18 |
| 10 | 369.72 | temperature | 25 | 1440 | > th | dose dependent | rect. | 11 | 23 |
| 36 | 370.18 | temperature | 25 | 1440 | > th | dose dependent | rect. | 14 | 23 |
| 54 | 370.38 | temperature | 25 | 1440 | > th | dose dependent | rect. | 21 | 22 |
| 78 | 370.61 | temperature | 25 | 1440 | > th | dose dependent | no | NA | NA |
| 86 | 370.68 | temperature | 25 | 1440 | > th | dose dependent | sin | 21 | NA |
| 109 | 371.07 | temperature | 25 | 1440 | > th | dose dependent | rect. | 8 | 23 |
| 182 | 372.48 | temperature | 20 | 1440 | > th | dose dependent | rect. | 17 | 2 |
| 209 | 373.37 | temperature | 20 | 1440 | > th | dose dependent | rect. | 16 | 4 |
| 219 | 373.60 | temperature | 20 | 1440 | > th | dose dependent | rect. | 15 | 7 |
| 263 | 375.16 | temperature | 10 | 720 | > th | dose dependent | rect. | 16 | 17 |
| 363 | 376.82 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 2 |
| 573 | 379.57 | temperature | 25 | 1440 | > th | dose dependent | rect. | 11 | 3 |
| 661 | 381.49 | temperature | 25 | 1440 | > th | dose dependent | rect. | 12 | 1 |