Os06g0175800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
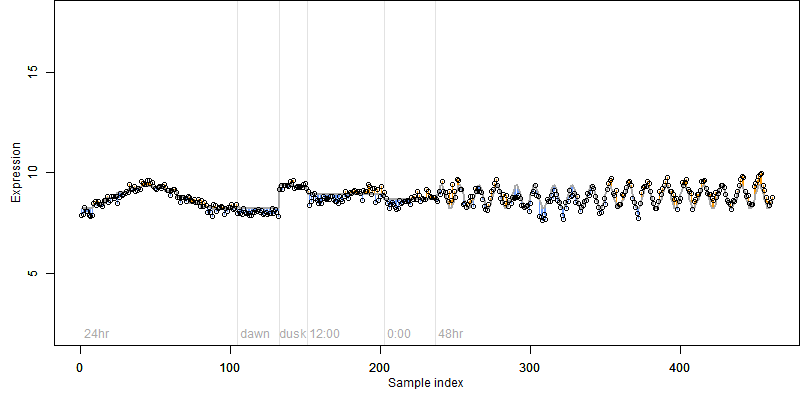
Description : Similar to Cystathionine beta-lyase, chloroplast precursor (EC 4.4.1.8) (CBL) (Beta-cystathionase) (Cysteine lyase).

FiT-DB / Search/ Help/ Sample detail
|
Os06g0175800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Cystathionine beta-lyase, chloroplast precursor (EC 4.4.1.8) (CBL) (Beta-cystathionase) (Cysteine lyase).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

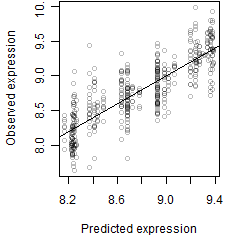
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22.87 | 0.225 | 8.73 | 0.886 | 1.18 | 0.3 | 0.474 | -1.11 | 0.226 | 17.4 | 23.8 | 843 | -- | -- |
| 25.41 | 0.235 | 8.77 | 0.559 | 1.16 | 0.406 | 0.467 | -- | 0.126 | 17.3 | 22.2 | 1987 | -- | -- |
| 23.29 | 0.225 | 8.73 | 0.866 | 1.19 | 0.298 | -- | -1.07 | 0.224 | 17.4 | 23.6 | 746 | -- | -- |
| 26.02 | 0.238 | 8.77 | 0.512 | 1.18 | 0.334 | -- | -- | 0.0901 | 17.2 | 22.1 | 1950 | -- | -- |
| 91.55 | 0.446 | 8.69 | 1.06 | -- | 0.491 | -- | -1.46 | 0.29 | -- | 24.2 | 1154 | -- | -- |
| 28.87 | 0.252 | 8.8 | 0.372 | 1.17 | -- | 0.58 | -- | -0.014 | 17.2 | -- | -- | -- | -- |
| 29.71 | 0.255 | 8.8 | 0.372 | 1.17 | -- | -- | -- | -0.0113 | 17.2 | -- | -- | -- | -- |
| 96.44 | 0.457 | 8.76 | 0.533 | -- | 0.419 | -- | -- | 0.0918 | -- | 21.5 | 2699 | -- | -- |
| 32.05 | 0.264 | 8.78 | -- | 1.13 | 0.26 | -- | -- | 0.0545 | 17.2 | 22.88 | 2080 | -- | -- |
| 101.72 | 0.471 | 8.78 | 0.331 | -- | -- | -- | -- | -0.0408 | -- | -- | -- | -- | -- |
| 33.36 | 0.27 | 8.81 | -- | 1.17 | -- | -- | -- | -0.0527 | 17.2 | -- | -- | -- | -- |
| 99.8 | 0.465 | 8.77 | -- | -- | 0.345 | -- | -- | 0.03 | -- | 24.05 | 2698 | -- | -- |
| 104.61 | 0.477 | 8.79 | -- | -- | -- | -- | -- | -0.0775 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21.27 | atmosphere | 1005 | 43200 | > th | dose independent | rect. | 5 | 2 |
| 4 | 21.30 | atmosphere | 1005 | 43200 | > th | dose independent | rect. | 4 | 5 |
| 7 | 21.31 | atmosphere | 1005 | 43200 | > th | dose independent | rect. | 4 | 8 |
| 8 | 21.31 | atmosphere | 1005 | 43200 | > th | dose independent | rect. | 2 | 10 |
| 9 | 21.31 | atmosphere | 1010 | 43200 | < th | dose dependent | rect. | 10 | 1 |
| 11 | 21.32 | atmosphere | 1005 | 43200 | > th | dose independent | rect. | 2 | 7 |
| 19 | 21.33 | atmosphere | 1005 | 43200 | < th | dose independent | rect. | 2 | 10 |
| 21 | 21.34 | atmosphere | 1005 | 43200 | < th | dose independent | rect. | 5 | 1 |
| 24 | 21.34 | atmosphere | 1005 | 43200 | < th | dose independent | rect. | 4 | 3 |
| 26 | 21.34 | atmosphere | 1005 | 43200 | < th | dose independent | rect. | 2 | 6 |
| 29 | 21.34 | atmosphere | 1005 | 43200 | < th | dose independent | rect. | 5 | 7 |
| 87 | 21.38 | atmosphere | 1005 | 43200 | > th | dose independent | rect. | 10 | 2 |
| 108 | 21.39 | atmosphere | 1005 | 43200 | < th | dose independent | rect. | 11 | 1 |
| 116 | 21.39 | atmosphere | 1005 | 43200 | > th | dose independent | rect. | 23 | 10 |
| 133 | 21.40 | atmosphere | 1005 | 43200 | > th | dose independent | rect. | 8 | 1 |
| 168 | 21.41 | atmosphere | 1005 | 43200 | > th | dose independent | sin | 6 | NA |
| 169 | 21.41 | atmosphere | 1005 | 43200 | < th | dose independent | sin | 5 | NA |
| 171 | 21.41 | wind | 5 | 43200 | > th | dose independent | rect. | 15 | 1 |
| 224 | 21.43 | wind | 5 | 43200 | < th | dose independent | rect. | 15 | 1 |
| 261 | 21.44 | atmosphere | 1010 | 43200 | < th | dose dependent | sin | 2 | NA |
| 589 | 21.54 | atmosphere | 1010 | 43200 | < th | dose dependent | no | NA | NA |
| 620 | 21.55 | atmosphere | 1010 | 43200 | < th | dose dependent | rect. | 13 | 1 |
| 640 | 21.56 | atmosphere | 1005 | 43200 | > th | dose independent | rect. | 2 | 1 |
| 657 | 21.56 | wind | 3 | 43200 | > th | dose dependent | rect. | 15 | 1 |
| 674 | 21.57 | atmosphere | 1005 | 43200 | > th | dose independent | rect. | 23 | 1 |
| 965 | 21.67 | atmosphere | 1010 | 43200 | < th | dose dependent | rect. | 23 | 1 |