Os06g0148300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.


FiT-DB / Search/ Help/ Sample detail
|
Os06g0148300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
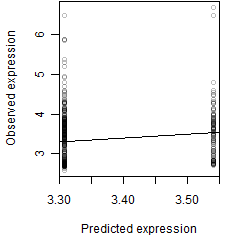
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

Residual plot

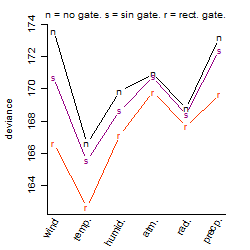
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 162.54 | 0.599 | 3.29 | 0.896 | 0.119 | -0.659 | 0.295 | -1.58 | 0.202 | 11.7 | 22.34 | 2020 | -- | -- |
| 168.63 | 0.605 | 3.35 | 0.401 | 0.144 | -0.535 | 0.404 | -- | 0.0572 | 11.1 | 23.4 | 909 | -- | -- |
| 162.9 | 0.594 | 3.29 | 0.903 | 0.112 | -0.652 | -- | -1.55 | 0.207 | 11.7 | 22.33 | 1961 | -- | -- |
| 167.79 | 0.603 | 3.35 | 0.365 | 0.0952 | -0.602 | -- | -- | 0.0442 | 11.5 | 23.4 | 2095 | -- | -- |
| 163.53 | 0.596 | 3.29 | 0.912 | -- | -0.63 | -- | -1.57 | 0.206 | -- | 22.45 | 1871 | -- | -- |
| 184.6 | 0.637 | 3.29 | 0.609 | 0.0828 | -- | 0.23 | -- | 0.301 | 12.7 | -- | -- | -- | -- |
| 184.83 | 0.637 | 3.29 | 0.617 | 0.0822 | -- | -- | -- | 0.302 | 12.5 | -- | -- | -- | -- |
| 168.7 | 0.605 | 3.36 | 0.348 | -- | -0.61 | -- | -- | 0.0383 | -- | 23.4 | 1974 | -- | -- |
| 170.3 | 0.608 | 3.37 | -- | 0.16 | -0.744 | -- | -- | -0.0593 | 11.8 | 23.33 | 2176 | -- | -- |
| 185.25 | 0.636 | 3.29 | 0.614 | -- | -- | -- | -- | 0.3 | -- | -- | -- | -- | -- |
| 194.87 | 0.653 | 3.31 | -- | 0.0734 | -- | -- | -- | 0.233 | 12.1 | -- | -- | -- | -- |
| 171.55 | 0.61 | 3.37 | -- | -- | -0.717 | -- | -- | -0.0486 | -- | 23.34 | 1974 | -- | -- |
| 195.21 | 0.652 | 3.31 | -- | -- | -- | -- | -- | 0.232 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 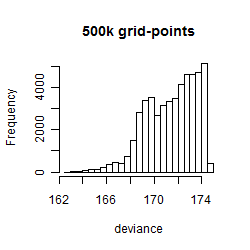
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 162.65 | temperature | 20 | 1440 | > th | dose dependent | rect. | 3 | 2 |
| 20 | 163.53 | temperature | 20 | 4320 | > th | dose dependent | rect. | 23 | 2 |
| 31 | 163.95 | temperature | 10 | 4320 | > th | dose dependent | rect. | 23 | 2 |
| 38 | 164.02 | temperature | 30 | 4320 | < th | dose dependent | rect. | 23 | 2 |
| 123 | 164.54 | temperature | 25 | 4320 | < th | dose dependent | rect. | 21 | 1 |
| 338 | 165.55 | temperature | 25 | 4320 | < th | dose dependent | sin | 13 | NA |
| 495 | 165.91 | temperature | 20 | 1440 | > th | dose dependent | sin | 10 | NA |
| 745 | 166.27 | temperature | 25 | 1440 | < th | dose dependent | rect. | 11 | 20 |
| 820 | 166.38 | temperature | 10 | 4320 | > th | dose dependent | sin | 11 | NA |
| 910 | 166.47 | temperature | 25 | 43200 | < th | dose independent | rect. | 19 | 2 |