Os06g0147200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.


FiT-DB / Search/ Help/ Sample detail
|
Os06g0147200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

Dependence on each variable

Residual plot
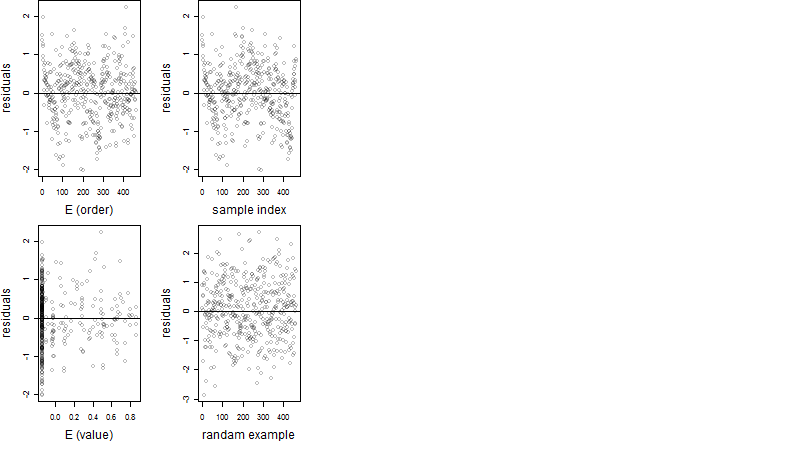
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 229.46 | 0.712 | 5.18 | -0.512 | 0.152 | -1.32 | 1.68 | 1.07 | -0.135 | 18.7 | 21.4 | 714 | 0.854 | -- |
| 230.66 | 0.707 | 5.15 | -0.34 | 0.194 | -1.41 | 1.33 | -- | -0.0788 | 17.3 | 21.3 | 720 | 0.159 | -- |
| 234.94 | 0.714 | 5.17 | -0.435 | 0.167 | -1.31 | -- | 0.597 | -0.109 | 18 | 21.4 | 758 | 0.997 | -- |
| 233.44 | 0.712 | 5.15 | -0.343 | 0.256 | -1.53 | -- | -- | -0.0954 | 15.8 | 21.33 | 739 | 0.000699 | -- |
| 236.98 | 0.717 | 5.17 | -0.381 | -- | -1.31 | -- | 0.399 | -0.119 | -- | 21.33 | 737 | 0.606 | -- |
| 286.26 | 0.793 | 5.09 | 0.0791 | 0.419 | -- | 0.718 | -- | 0.23 | 19.8 | -- | -- | -- | -- |
| 287.13 | 0.794 | 5.09 | 0.0562 | 0.422 | -- | -- | -- | 0.229 | 20.7 | -- | -- | -- | -- |
| 236.63 | 0.716 | 5.15 | -0.331 | -- | -1.47 | -- | -- | -0.0839 | -- | 21.33 | 693 | 2.28e-05 | -- |
| 236.23 | 0.716 | 5.14 | -- | 0.25 | -1.38 | -- | -- | -0.0456 | 15.9 | 21.64 | 714 | 2.25e-06 | -- |
| 297.22 | 0.806 | 5.08 | 0.0553 | -- | -- | -- | -- | 0.225 | -- | -- | -- | -- | -- |
| 287.22 | 0.793 | 5.09 | -- | 0.422 | -- | -- | -- | 0.222 | 20.7 | -- | -- | -- | -- |
| 239.32 | 0.721 | 5.14 | -- | -- | -1.4 | -- | -- | -0.0326 | -- | 21.4 | 685 | 6.77e-06 | -- |
| 297.3 | 0.805 | 5.08 | -- | -- | -- | -- | -- | 0.219 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance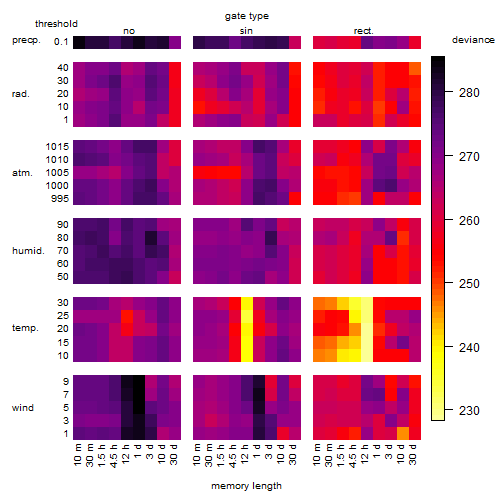
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 228.27 | temperature | 15 | 720 | > th | dose dependent | rect. | 1 | 8 |
| 3 | 228.96 | temperature | 20 | 720 | > th | dose dependent | rect. | 6 | 1 |
| 4 | 229.26 | temperature | 20 | 720 | > th | dose dependent | rect. | 3 | 6 |
| 12 | 229.91 | temperature | 15 | 720 | > th | dose dependent | rect. | 23 | 10 |
| 18 | 230.61 | temperature | 25 | 720 | < th | dose dependent | rect. | 5 | 2 |
| 19 | 230.69 | temperature | 30 | 720 | < th | dose dependent | rect. | 2 | 7 |
| 21 | 230.72 | temperature | 25 | 720 | < th | dose dependent | rect. | 4 | 4 |
| 52 | 232.21 | temperature | 25 | 720 | < th | dose dependent | rect. | 22 | 10 |
| 118 | 234.31 | temperature | 20 | 720 | < th | dose independent | rect. | 3 | 4 |
| 124 | 234.53 | temperature | 25 | 720 | < th | dose dependent | sin | 7 | NA |
| 143 | 234.96 | temperature | 20 | 720 | > th | dose independent | rect. | 3 | 4 |
| 156 | 235.26 | temperature | 20 | 720 | > th | dose independent | rect. | 1 | 6 |
| 164 | 235.41 | temperature | 30 | 270 | < th | dose dependent | rect. | 7 | 7 |
| 196 | 236.11 | temperature | 20 | 720 | < th | dose independent | rect. | 23 | 8 |
| 205 | 236.31 | temperature | 20 | 720 | > th | dose independent | rect. | 23 | 8 |
| 309 | 238.18 | temperature | 15 | 720 | > th | dose dependent | sin | 7 | NA |
| 366 | 239.33 | temperature | 10 | 270 | > th | dose dependent | rect. | 4 | 14 |
| 371 | 239.40 | temperature | 20 | 720 | < th | dose independent | sin | 7 | NA |
| 380 | 239.62 | temperature | 25 | 270 | < th | dose independent | rect. | 7 | 6 |
| 404 | 240.10 | temperature | 30 | 270 | < th | dose dependent | rect. | 9 | 2 |
| 573 | 242.62 | temperature | 20 | 720 | > th | dose independent | sin | 7 | NA |
| 766 | 245.33 | temperature | 20 | 720 | > th | dose independent | rect. | 1 | 16 |
| 810 | 245.76 | wind | 1 | 14400 | < th | dose dependent | rect. | 24 | 2 |
| 847 | 246.05 | temperature | 20 | 720 | > th | dose independent | rect. | 3 | 14 |