Os06g0113800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Kinetechore (Skp1p-like) protein-like.


FiT-DB / Search/ Help/ Sample detail
|
Os06g0113800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Kinetechore (Skp1p-like) protein-like.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

Residual plot

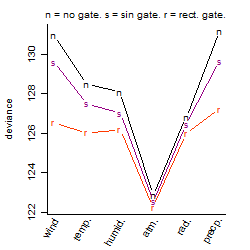
Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 126.41 | 0.528 | 3.85 | 0.286 | 0.224 | 0.529 | 0.862 | 1.43 | -0.026 | 17.4 | 1011 | 10214 | -- | -- |
| 127.97 | 0.527 | 3.82 | 0.877 | 0.273 | 0.603 | 0.932 | -- | 0.0721 | 18.1 | 1012 | 20151 | -- | -- |
| 126.53 | 0.524 | 3.83 | 0.419 | 0.251 | 0.65 | -- | 1.26 | 0.0386 | 17.6 | 1012 | 17073 | -- | -- |
| 129.64 | 0.53 | 3.8 | 0.889 | 0.254 | 0.611 | -- | -- | 0.114 | 18.1 | 1012 | 20241 | -- | -- |
| 129.33 | 0.53 | 3.83 | 0.461 | -- | 0.624 | -- | 1.18 | 0.00937 | -- | 1012 | 17259 | -- | -- |
| 144.24 | 0.563 | 3.84 | 0.623 | 0.254 | -- | 0.977 | -- | -0.0772 | 17.2 | -- | -- | -- | -- |
| 146.52 | 0.567 | 3.84 | 0.622 | 0.265 | -- | -- | -- | -0.0729 | 17.8 | -- | -- | -- | -- |
| 133.05 | 0.537 | 3.8 | 0.88 | -- | 0.618 | -- | -- | 0.111 | -- | 1012 | 20172 | -- | -- |
| 147.9 | 0.566 | 3.83 | -- | 0.24 | 0.533 | -- | -- | -0.00521 | 18.3 | 1011 | 16951 | -- | -- |
| 150.17 | 0.573 | 3.84 | 0.614 | -- | -- | -- | -- | -0.0791 | -- | -- | -- | -- | -- |
| 156.71 | 0.586 | 3.86 | -- | 0.256 | -- | -- | -- | -0.142 | 18 | -- | -- | -- | -- |
| 150.73 | 0.572 | 3.83 | -- | -- | 0.473 | -- | -- | -0.0549 | -- | 1012 | 19554 | -- | -- |
| 160.11 | 0.591 | 3.85 | -- | -- | -- | -- | -- | -0.147 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 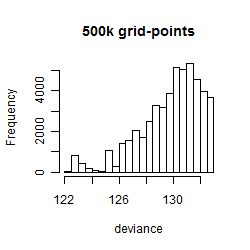
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 122.26 | atmosphere | 1015 | 43200 | < th | dose independent | rect. | 24 | 1 |
| 2 | 122.37 | atmosphere | 1015 | 43200 | > th | dose independent | rect. | 24 | 1 |
| 4 | 122.41 | atmosphere | 1015 | 43200 | < th | dose independent | rect. | 4 | 1 |
| 5 | 122.42 | atmosphere | 1015 | 43200 | > th | dose independent | rect. | 3 | 1 |
| 7 | 122.43 | atmosphere | 1015 | 43200 | < th | dose independent | rect. | 22 | 7 |
| 11 | 122.47 | atmosphere | 1015 | 43200 | > th | dose independent | rect. | 18 | 11 |
| 12 | 122.48 | atmosphere | 1015 | 43200 | > th | dose independent | rect. | 20 | 1 |
| 21 | 122.49 | atmosphere | 1015 | 43200 | < th | dose independent | rect. | 18 | 11 |
| 31 | 122.51 | atmosphere | 1015 | 43200 | > th | dose independent | rect. | 22 | 7 |
| 59 | 122.55 | atmosphere | 1015 | 43200 | > th | dose independent | sin | 13 | NA |
| 85 | 122.57 | atmosphere | 1015 | 43200 | < th | dose independent | sin | 12 | NA |
| 169 | 122.64 | atmosphere | 1015 | 43200 | > th | dose dependent | rect. | 21 | 10 |
| 189 | 122.66 | atmosphere | 1015 | 43200 | > th | dose dependent | rect. | 23 | 7 |
| 240 | 122.71 | atmosphere | 1015 | 43200 | > th | dose dependent | sin | 9 | NA |
| 305 | 122.76 | atmosphere | 1015 | 43200 | > th | dose dependent | rect. | 1 | 4 |
| 494 | 122.86 | atmosphere | 1015 | 43200 | < th | dose independent | no | NA | NA |
| 526 | 122.87 | atmosphere | 1015 | 43200 | > th | dose independent | no | NA | NA |
| 531 | 122.87 | atmosphere | 1015 | 43200 | > th | dose independent | rect. | 3 | 22 |
| 535 | 122.87 | atmosphere | 1015 | 43200 | < th | dose independent | rect. | 2 | 23 |
| 553 | 122.88 | atmosphere | 1015 | 43200 | < th | dose independent | rect. | 21 | 23 |
| 606 | 122.90 | atmosphere | 1015 | 43200 | > th | dose dependent | rect. | 2 | 2 |
| 697 | 122.94 | atmosphere | 1015 | 43200 | < th | dose independent | rect. | 19 | 2 |
| 712 | 122.94 | atmosphere | 1015 | 43200 | > th | dose dependent | no | NA | NA |
| 824 | 123.00 | atmosphere | 1015 | 43200 | > th | dose dependent | rect. | 9 | 23 |
| 917 | 123.08 | atmosphere | 1015 | 43200 | > th | dose dependent | rect. | 5 | 1 |