Os05g0518300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
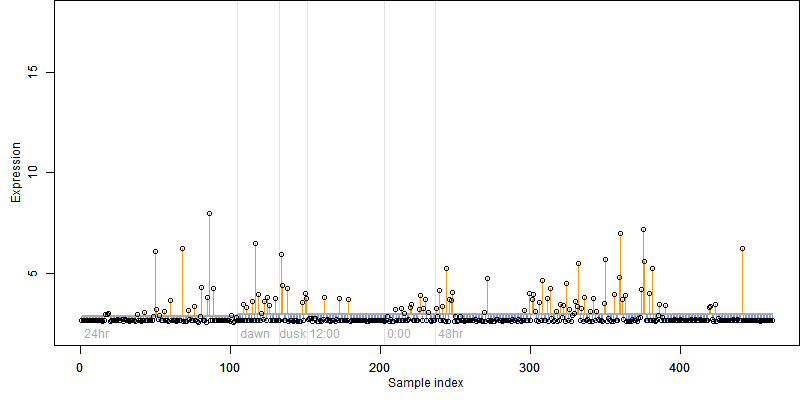
Description : Lipolytic enzyme, G-D-S-L family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os05g0518300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Lipolytic enzyme, G-D-S-L family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
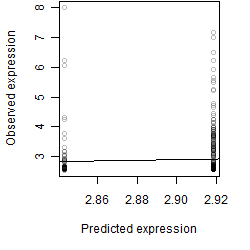
Dependence on each variable

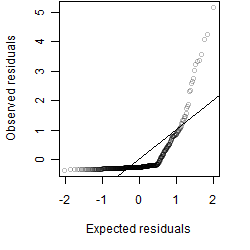
Residual plot

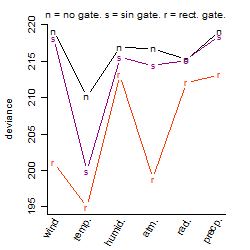
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 209.59 | 0.68 | 2.85 | 0.446 | 0.235 | -0.938 | -0.228 | 18.8 | -0.0486 | 3.43 | 30.65 | 30 | -- | -- |
| 213.45 | 0.686 | 2.91 | -0.0897 | 0.235 | 1.24 | -0.264 | -- | -0.0459 | 3.34 | 30.65 | 30 | -- | -- |
| 209.73 | 0.675 | 2.85 | 0.446 | 0.233 | -0.952 | -- | 19.1 | -0.0466 | 3.21 | 30.71 | 30 | -- | -- |
| 212.98 | 0.68 | 2.91 | -0.0901 | 0.245 | 1.3 | -- | -- | -0.0405 | 3.29 | 30.6 | 30 | -- | -- |
| 212.58 | 0.679 | 2.85 | 0.473 | -- | -1.26 | -- | 20.4 | -0.0481 | -- | 30.84 | 30 | -- | -- |
| 224.25 | 0.702 | 2.92 | -0.0324 | 0.113 | -- | -0.583 | -- | -0.0837 | 4.16 | -- | -- | -- | -- |
| 225.08 | 0.703 | 2.92 | -0.0266 | 0.114 | -- | -- | -- | -0.0804 | 2.62 | -- | -- | -- | -- |
| 216.09 | 0.685 | 2.91 | -0.0687 | -- | 1.16 | -- | -- | -0.0421 | -- | 30.8 | 30 | -- | -- |
| 213.19 | 0.68 | 2.91 | -- | 0.241 | 1.28 | -- | -- | -0.0312 | 3.29 | 30.6 | 30 | -- | -- |
| 225.85 | 0.702 | 2.92 | -0.0218 | -- | -- | -- | -- | -0.0775 | -- | -- | -- | -- | -- |
| 225.1 | 0.702 | 2.92 | -- | 0.114 | -- | -- | -- | -0.0775 | 2.62 | -- | -- | -- | -- |
| 216.22 | 0.685 | 2.91 | -- | -- | 1.14 | -- | -- | -0.0341 | -- | 30.8 | 30 | -- | -- |
| 225.86 | 0.701 | 2.92 | -- | -- | -- | -- | -- | -0.0751 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance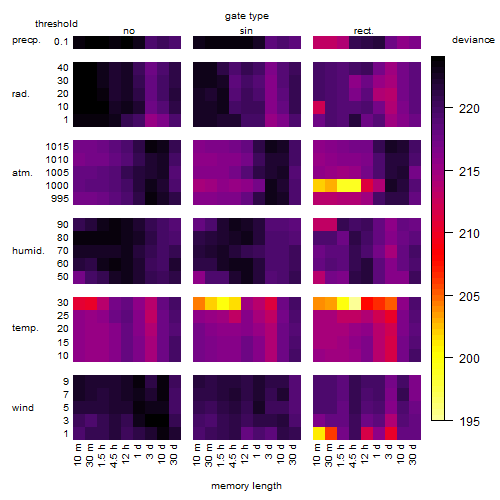
|
Histogram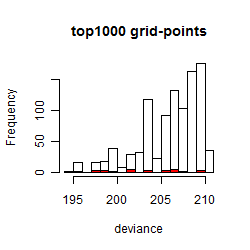 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 194.95 | temperature | 30 | 270 | > th | dose dependent | rect. | 18 | 17 |
| 2 | 195.09 | temperature | 30 | 270 | > th | dose dependent | rect. | 8 | 3 |
| 17 | 197.49 | temperature | 30 | 270 | > th | dose independent | rect. | 18 | 17 |
| 18 | 197.78 | temperature | 30 | 270 | > th | dose independent | rect. | 8 | 3 |
| 33 | 198.78 | atmosphere | 1000 | 270 | < th | dose independent | rect. | 9 | 2 |
| 48 | 198.97 | atmosphere | 1000 | 90 | < th | dose dependent | rect. | 11 | 2 |
| 49 | 198.98 | atmosphere | 1000 | 270 | < th | dose dependent | rect. | 9 | 3 |
| 85 | 199.86 | temperature | 30 | 90 | > th | dose dependent | sin | 8 | NA |
| 96 | 201.10 | wind | 1 | 10 | < th | dose dependent | rect. | 11 | 1 |
| 120 | 201.52 | temperature | 30 | 270 | > th | dose independent | sin | 9 | NA |
| 122 | 201.81 | wind | 1 | 10 | < th | dose independent | rect. | 11 | 2 |
| 124 | 201.98 | atmosphere | 1000 | 10 | < th | dose dependent | rect. | 10 | 4 |
| 225 | 203.47 | temperature | 30 | 270 | < th | dose independent | sin | 9 | NA |
| 252 | 203.71 | wind | 1 | 10 | < th | dose dependent | rect. | 11 | 7 |
| 260 | 203.72 | temperature | 30 | 10 | > th | dose dependent | rect. | 17 | 20 |
| 308 | 205.35 | temperature | 30 | 4320 | < th | dose independent | rect. | 3 | 6 |
| 355 | 205.75 | temperature | 30 | 4320 | > th | dose independent | rect. | 8 | 1 |
| 390 | 206.05 | temperature | 30 | 4320 | > th | dose dependent | rect. | 8 | 1 |
| 434 | 206.30 | temperature | 30 | 4320 | < th | dose independent | rect. | 23 | 10 |
| 438 | 206.38 | temperature | 30 | 4320 | < th | dose independent | rect. | 19 | 14 |
| 479 | 206.70 | temperature | 30 | 4320 | < th | dose independent | rect. | 8 | 1 |
| 564 | 207.62 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 10 | 2 |
| 851 | 209.49 | temperature | 30 | 30 | > th | dose dependent | rect. | 9 | 9 |
| 964 | 210.00 | wind | 1 | 4320 | > th | dose independent | rect. | 2 | 1 |