Os05g0166300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Curculin-like (mannose-binding) lectin domain containing protein.
FiT-DB / Search/ Help/ Sample detail
|
Os05g0166300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Curculin-like (mannose-binding) lectin domain containing protein.
|
|
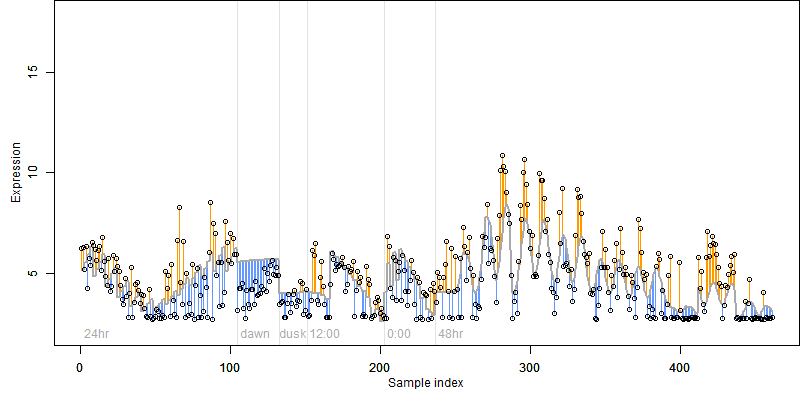
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

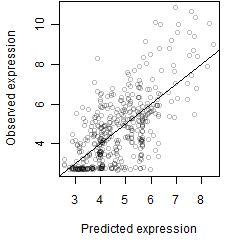
Residual plot
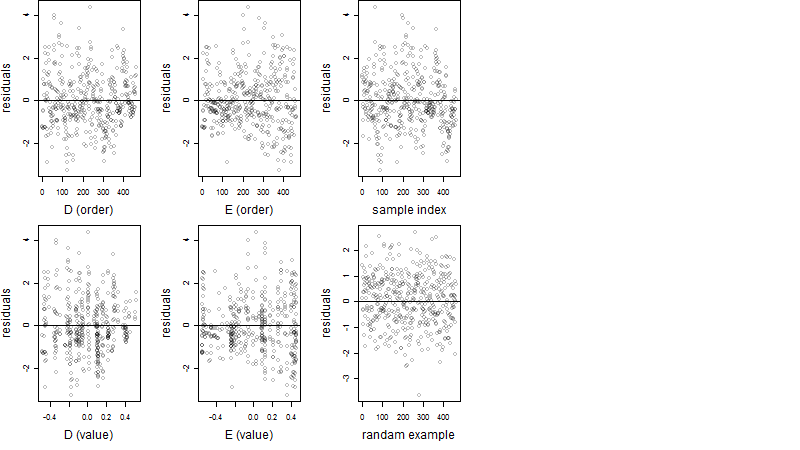
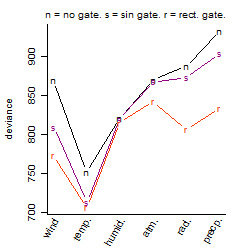
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 709.79 | 1.25 | 4.96 | -4.02 | 0.6 | 3.33 | 1.84 | -4.04 | -1.13 | 14.5 | 20.2 | 758 | 7.09 | -- |
| 740.3 | 1.27 | 4.9 | -3.5 | 0.977 | 3.67 | 3.25 | -- | -0.99 | 14.8 | 20.22 | 774 | 7.52 | -- |
| 716.23 | 1.25 | 4.97 | -4.12 | 0.305 | 3.13 | -- | -4.84 | -1.15 | 14.4 | 20.4 | 801 | 7.12 | -- |
| 781.39 | 1.3 | 4.88 | -3.35 | 0.0607 | 3.13 | -- | -- | -1 | 2.59 | 20.69 | 897 | 7.1 | -- |
| 718.36 | 1.25 | 5.01 | -4.21 | -- | 3.04 | -- | -5 | -1.19 | -- | 20.5 | 833 | 7.11 | -- |
| 983.8 | 1.47 | 4.72 | -2.47 | 1.76 | -- | -3.78 | -- | -0.393 | 4.94 | -- | -- | -- | -- |
| 1017.14 | 1.49 | 4.7 | -2.45 | 1.76 | -- | -- | -- | -0.373 | 5.45 | -- | -- | -- | -- |
| 781.38 | 1.3 | 4.88 | -3.35 | -- | 3.16 | -- | -- | -1 | -- | 20.7 | 894 | 7.2 | -- |
| 1045.52 | 1.51 | 4.76 | -- | 0.233 | 2.41 | -- | -- | -0.49 | 1.09 | 20.66 | 750 | 5.65 | -- |
| 1178.62 | 1.6 | 4.72 | -2.39 | -- | -- | -- | -- | -0.33 | -- | -- | -- | -- | -- |
| 1175.65 | 1.6 | 4.64 | -- | 1.72 | -- | -- | -- | -0.1 | 5.51 | -- | -- | -- | -- |
| 1046.35 | 1.51 | 4.76 | -- | -- | 2.48 | -- | -- | -0.496 | -- | 20.61 | 737 | 6.02 | -- |
| 1329.74 | 1.7 | 4.66 | -- | -- | -- | -- | -- | -0.0644 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance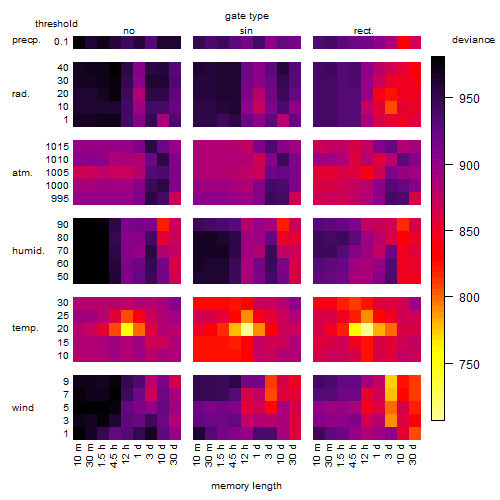
|
Histogram 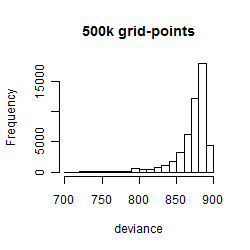
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 706.80 | temperature | 20 | 720 | > th | dose independent | rect. | 16 | 15 |
| 6 | 713.33 | temperature | 20 | 720 | > th | dose independent | sin | 13 | NA |
| 22 | 724.48 | temperature | 20 | 720 | < th | dose independent | rect. | 16 | 15 |
| 24 | 725.02 | temperature | 20 | 720 | < th | dose independent | sin | 12 | NA |
| 48 | 728.68 | temperature | 20 | 720 | > th | dose independent | rect. | 18 | 21 |
| 49 | 728.78 | temperature | 20 | 720 | < th | dose independent | rect. | 19 | 10 |
| 243 | 751.39 | temperature | 20 | 720 | > th | dose independent | no | NA | NA |
| 269 | 753.85 | temperature | 20 | 720 | < th | dose independent | no | NA | NA |
| 280 | 754.76 | temperature | 20 | 720 | > th | dose independent | rect. | 1 | 23 |
| 287 | 755.76 | temperature | 20 | 720 | > th | dose independent | rect. | 3 | 23 |
| 316 | 759.01 | temperature | 20 | 720 | < th | dose independent | rect. | 3 | 23 |
| 324 | 759.99 | temperature | 20 | 720 | < th | dose independent | rect. | 1 | 23 |
| 343 | 762.92 | temperature | 20 | 720 | < th | dose independent | rect. | 23 | 2 |
| 409 | 772.78 | wind | 9 | 4320 | > th | dose independent | rect. | 17 | 5 |
| 410 | 773.18 | wind | 9 | 4320 | < th | dose independent | rect. | 17 | 4 |
| 427 | 775.84 | wind | 7 | 4320 | > th | dose dependent | rect. | 18 | 3 |
| 449 | 778.30 | wind | 9 | 4320 | > th | dose independent | rect. | 17 | 11 |
| 450 | 778.34 | wind | 7 | 4320 | < th | dose independent | rect. | 18 | 2 |
| 454 | 778.52 | wind | 7 | 4320 | > th | dose dependent | rect. | 17 | 5 |
| 466 | 779.95 | wind | 7 | 4320 | > th | dose independent | rect. | 18 | 1 |
| 550 | 787.56 | wind | 9 | 4320 | < th | dose independent | rect. | 17 | 11 |
| 553 | 787.61 | wind | 9 | 4320 | < th | dose independent | rect. | 17 | 9 |
| 559 | 787.88 | wind | 7 | 4320 | > th | dose dependent | rect. | 17 | 8 |
| 609 | 789.78 | temperature | 25 | 720 | < th | dose dependent | rect. | 16 | 18 |
| 686 | 792.44 | wind | 3 | 4320 | > th | dose dependent | rect. | 24 | 1 |
| 693 | 792.57 | temperature | 25 | 720 | < th | dose dependent | sin | 11 | NA |
| 770 | 793.82 | wind | 9 | 4320 | < th | dose independent | rect. | 17 | 16 |