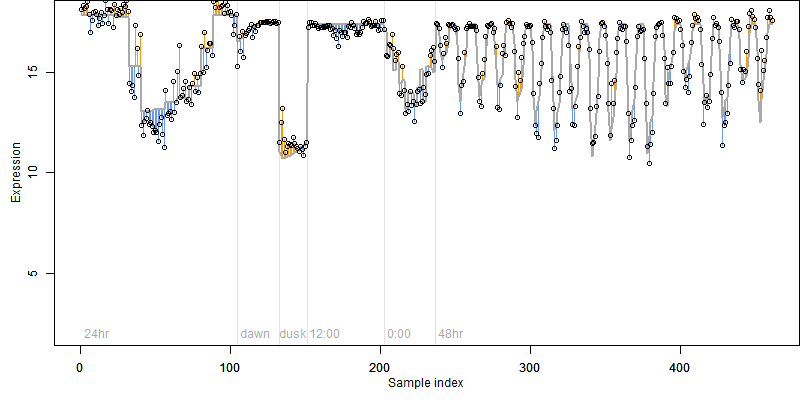
Os04g0683700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to 4-coumarate-CoA ligase-like protein (Adenosine monophosphate binding protein 3 AMPBP3).

FiT-DB / Search/ Help/ Sample detail
|
Os04g0683700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to 4-coumarate-CoA ligase-like protein (Adenosine monophosphate binding protein 3 AMPBP3).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
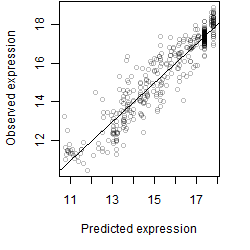
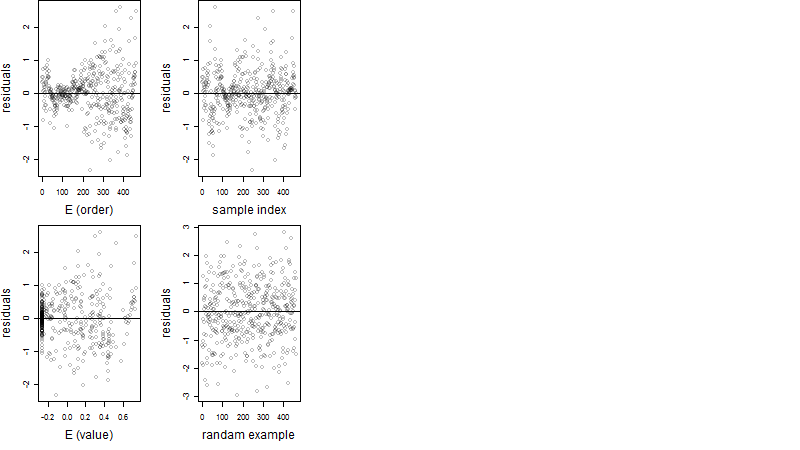
Dependence on each variable

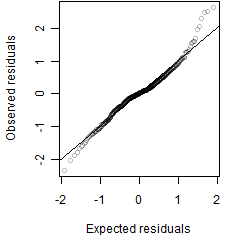
Residual plot
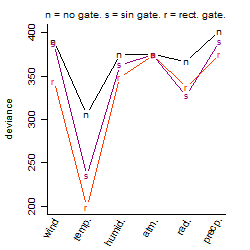
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 193.36 | 0.653 | 15.6 | 0.282 | 1.15 | -5.86 | 1.98 | 3.94 | 0.476 | 7.67 | 15.58 | 264 | 13.1 | 11.4 |
| 200.36 | 0.659 | 15.6 | 0.0772 | 1.45 | -5.54 | 0.596 | -- | 0.457 | 7.68 | 16.2 | 241 | 13.2 | 11.8 |
| 196.49 | 0.653 | 15.6 | 0.112 | 1.15 | -5.82 | -- | 1.4 | 0.395 | 7.27 | 15.5 | 268 | 13.1 | 11.5 |
| 197.96 | 0.655 | 15.6 | 0.0134 | 1.09 | -5.85 | -- | -- | 0.41 | 7.14 | 15.16 | 256 | 13.2 | 11.5 |
| 209.4 | 0.674 | 15.6 | 0.388 | -- | -6.73 | -- | 1.57 | 0.538 | -- | 13.6 | 243 | 13.1 | 11.7 |
| 480.84 | 1.03 | 15.6 | -0.705 | 5.28 | -- | 1.87 | -- | 0.0121 | 7.96 | -- | -- | -- | -- |
| 491.13 | 1.04 | 15.6 | -0.655 | 5.28 | -- | -- | -- | 0.01 | 7.95 | -- | -- | -- | -- |
| 208.23 | 0.672 | 15.6 | 0.137 | -- | -6.67 | -- | -- | 0.504 | -- | 13.59 | 250 | 12.9 | 11.7 |
| 197.92 | 0.655 | 15.6 | -- | 1.08 | -5.84 | -- | -- | 0.395 | 7.13 | 15.11 | 256 | 13.2 | 11.5 |
| 2010.01 | 2.09 | 15.7 | -0.6 | -- | -- | -- | -- | 0.0789 | -- | -- | -- | -- | -- |
| 502.43 | 1.05 | 15.6 | -- | 5.27 | -- | -- | -- | 0.083 | 7.96 | -- | -- | -- | -- |
| 206.89 | 0.67 | 15.7 | -- | -- | -6.67 | -- | -- | 0.44 | -- | 13.45 | 258 | 12.9 | 11.6 |
| 2019.51 | 2.1 | 15.7 | -- | -- | -- | -- | -- | 0.146 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance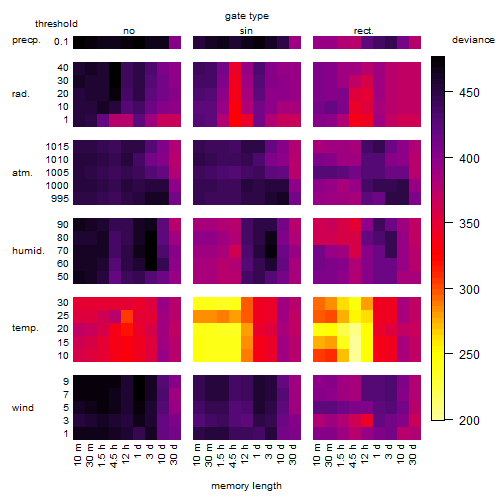
|
Histogram 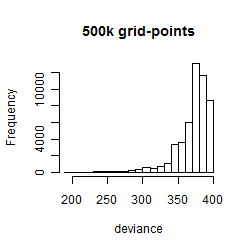
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 198.92 | temperature | 15 | 270 | > th | dose dependent | rect. | 13 | 11 |
| 17 | 227.93 | temperature | 20 | 270 | > th | dose dependent | rect. | 13 | 19 |
| 28 | 234.97 | temperature | 20 | 270 | > th | dose dependent | sin | 16 | NA |
| 31 | 235.76 | temperature | 20 | 90 | > th | dose dependent | sin | 14 | NA |
| 37 | 236.84 | temperature | 30 | 90 | < th | dose dependent | sin | 15 | NA |
| 42 | 238.27 | temperature | 30 | 270 | < th | dose dependent | sin | 17 | NA |
| 69 | 243.23 | temperature | 30 | 270 | < th | dose dependent | rect. | 11 | 16 |
| 90 | 246.54 | temperature | 20 | 10 | > th | dose dependent | rect. | 16 | 11 |
| 103 | 248.05 | temperature | 20 | 720 | > th | dose dependent | rect. | 13 | 4 |
| 148 | 255.02 | temperature | 25 | 270 | > th | dose independent | rect. | 13 | 11 |
| 183 | 260.98 | temperature | 30 | 270 | < th | dose dependent | rect. | 15 | 12 |
| 233 | 265.95 | temperature | 30 | 90 | < th | dose dependent | rect. | 17 | 12 |
| 260 | 269.23 | temperature | 20 | 10 | > th | dose dependent | rect. | 16 | 18 |
| 322 | 274.47 | temperature | 25 | 90 | > th | dose independent | rect. | 15 | 12 |
| 355 | 277.62 | temperature | 25 | 270 | > th | dose independent | rect. | 13 | 19 |
| 359 | 277.84 | temperature | 10 | 720 | > th | dose dependent | rect. | 6 | 19 |
| 382 | 279.04 | temperature | 20 | 720 | > th | dose dependent | sin | 21 | NA |
| 437 | 282.09 | temperature | 30 | 720 | < th | dose dependent | rect. | 10 | 10 |
| 465 | 283.43 | temperature | 20 | 720 | > th | dose dependent | rect. | 6 | 18 |
| 578 | 287.80 | temperature | 25 | 270 | > th | dose independent | sin | 18 | NA |
| 601 | 288.62 | temperature | 25 | 270 | < th | dose independent | sin | 18 | NA |
| 631 | 290.05 | temperature | 20 | 720 | > th | dose dependent | rect. | 6 | 13 |
| 633 | 290.18 | temperature | 10 | 720 | > th | dose dependent | rect. | 5 | 14 |
| 678 | 291.58 | temperature | 25 | 270 | < th | dose independent | rect. | 10 | 15 |
| 971 | 299.62 | temperature | 25 | 720 | > th | dose dependent | rect. | 21 | 21 |