Os04g0630300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : NAD-dependent epimerase/dehydratase family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os04g0630300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : NAD-dependent epimerase/dehydratase family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

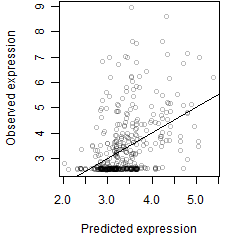 __
__
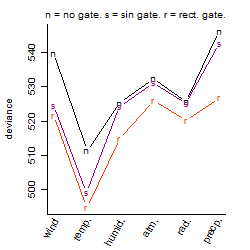
Dependence on each variable

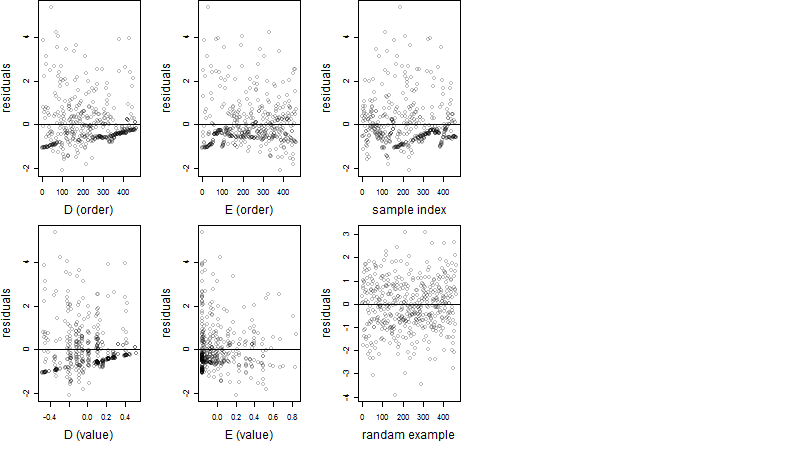
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 486.65 | 1.04 | 3.54 | -2.64 | 0.683 | 2.06 | -2.02 | -9.15 | -0.653 | 18 | 22.6 | 40 | 0.484 | -- |
| 529.07 | 1.07 | 3.41 | -1.49 | 0.521 | 1.38 | -1.62 | -- | -0.269 | 16.3 | 22.81 | 40 | 0.563 | -- |
| 494.68 | 1.04 | 3.52 | -2.48 | 0.547 | 2.02 | -- | -9.17 | -0.604 | 17.6 | 23.3 | 38 | 0.486 | -- |
| 535.19 | 1.08 | 3.42 | -1.55 | 0.646 | 1.51 | -- | -- | -0.294 | 17.6 | 22.12 | 39 | 0.155 | -- |
| 506.79 | 1.05 | 3.52 | -2.31 | -- | 1.81 | -- | -8.94 | -0.575 | -- | 23.6 | 38 | 0.00605 | -- |
| 559.5 | 1.11 | 3.34 | -1.29 | 0.491 | -- | -1.22 | -- | -0.0312 | 13.8 | -- | -- | -- | -- |
| 564.96 | 1.11 | 3.34 | -1.33 | 0.491 | -- | -- | -- | -0.0395 | 13.2 | -- | -- | -- | -- |
| 545.35 | 1.09 | 3.39 | -1.49 | -- | 1.3 | -- | -- | -0.232 | -- | 23.85 | 26 | 6.95e-06 | -- |
| 594.78 | 1.14 | 3.35 | -- | 0.414 | 1.11 | -- | -- | -0.0395 | 16.7 | 23.3 | 36 | 0.353 | -- |
| 579.9 | 1.13 | 3.35 | -1.35 | -- | -- | -- | -- | -0.0493 | -- | -- | -- | -- | -- |
| 611.87 | 1.16 | 3.31 | -- | 0.514 | -- | -- | -- | 0.109 | 13.3 | -- | -- | -- | -- |
| 602.62 | 1.15 | 3.35 | -- | -- | 1.11 | -- | -- | -0.0385 | -- | 23.99 | 26 | 2.59e-07 | -- |
| 628.18 | 1.17 | 3.32 | -- | -- | -- | -- | -- | 0.101 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 494.72 | temperature | 25 | 30 | < th | dose independent | rect. | 19 | 22 |
| 8 | 496.85 | temperature | 25 | 30 | < th | dose independent | rect. | 5 | 12 |
| 16 | 498.29 | temperature | 25 | 90 | < th | dose independent | rect. | 23 | 17 |
| 19 | 498.94 | temperature | 20 | 30 | > th | dose dependent | sin | 5 | NA |
| 23 | 499.62 | temperature | 25 | 30 | > th | dose independent | sin | 4 | NA |
| 44 | 502.46 | temperature | 25 | 90 | < th | dose independent | sin | 5 | NA |
| 66 | 504.56 | temperature | 25 | 10 | > th | dose independent | rect. | 7 | 5 |
| 67 | 504.56 | temperature | 25 | 30 | > th | dose independent | rect. | 3 | 9 |
| 73 | 504.80 | temperature | 30 | 10 | < th | dose dependent | rect. | 18 | 23 |
| 80 | 505.55 | temperature | 20 | 30 | > th | dose dependent | rect. | 21 | 15 |
| 91 | 506.26 | temperature | 30 | 10 | < th | dose dependent | sin | 5 | NA |
| 99 | 506.60 | temperature | 20 | 30 | > th | dose dependent | rect. | 1 | 11 |
| 116 | 507.06 | temperature | 25 | 270 | > th | dose dependent | rect. | 24 | 9 |
| 136 | 507.99 | temperature | 25 | 270 | > th | dose dependent | rect. | 7 | 1 |
| 137 | 508.06 | temperature | 25 | 270 | > th | dose dependent | rect. | 22 | 11 |
| 143 | 508.20 | temperature | 20 | 10 | > th | dose dependent | rect. | 6 | 23 |
| 149 | 508.55 | temperature | 25 | 10 | > th | dose independent | rect. | 3 | 13 |
| 189 | 509.67 | temperature | 25 | 270 | > th | dose dependent | rect. | 6 | 3 |
| 197 | 509.80 | temperature | 20 | 10 | > th | dose dependent | rect. | 17 | 23 |
| 212 | 510.44 | temperature | 20 | 90 | > th | dose dependent | rect. | 13 | 23 |
| 224 | 510.80 | temperature | 20 | 10 | > th | dose dependent | rect. | 21 | 23 |
| 235 | 511.02 | temperature | 30 | 10 | < th | dose dependent | rect. | 5 | 12 |
| 249 | 511.23 | temperature | 20 | 10 | > th | dose dependent | no | NA | NA |
| 250 | 511.26 | temperature | 25 | 10 | > th | dose independent | rect. | 22 | 18 |
| 251 | 511.27 | temperature | 25 | 10 | > th | dose independent | rect. | 22 | 14 |
| 261 | 511.39 | temperature | 30 | 90 | < th | dose dependent | rect. | 23 | 17 |
| 262 | 511.40 | temperature | 20 | 10 | > th | dose dependent | rect. | 1 | 23 |
| 281 | 511.65 | temperature | 20 | 10 | > th | dose dependent | rect. | 4 | 23 |
| 284 | 511.67 | temperature | 20 | 10 | > th | dose dependent | rect. | 8 | 23 |
| 288 | 511.70 | temperature | 20 | 90 | > th | dose dependent | rect. | 15 | 23 |
| 294 | 511.91 | temperature | 25 | 10 | < th | dose independent | rect. | 22 | 23 |
| 340 | 512.65 | temperature | 20 | 10 | > th | dose dependent | rect. | 15 | 23 |
| 399 | 513.51 | temperature | 30 | 10 | < th | dose dependent | rect. | 23 | 22 |
| 436 | 514.12 | temperature | 30 | 90 | < th | dose dependent | rect. | 23 | 22 |
| 447 | 514.29 | temperature | 25 | 10 | > th | dose independent | rect. | 19 | 21 |
| 494 | 514.91 | humidity | 50 | 43200 | < th | dose independent | rect. | 16 | 2 |
| 519 | 515.19 | temperature | 20 | 10 | > th | dose dependent | rect. | 6 | 18 |
| 524 | 515.38 | temperature | 20 | 30 | > th | dose dependent | rect. | 17 | 19 |
| 536 | 515.46 | temperature | 30 | 90 | < th | dose dependent | rect. | 11 | 23 |
| 537 | 515.47 | humidity | 50 | 43200 | > th | dose independent | rect. | 16 | 2 |
| 570 | 515.79 | temperature | 25 | 270 | > th | dose independent | rect. | 6 | 3 |
| 589 | 515.99 | temperature | 30 | 30 | < th | dose dependent | rect. | 9 | 23 |
| 638 | 516.53 | temperature | 30 | 10 | < th | dose dependent | rect. | 23 | 18 |
| 641 | 516.57 | temperature | 20 | 10 | > th | dose dependent | rect. | 11 | 23 |
| 648 | 516.62 | temperature | 30 | 10 | < th | dose dependent | rect. | 13 | 23 |
| 664 | 516.76 | humidity | 50 | 43200 | > th | dose independent | rect. | 16 | 8 |
| 668 | 516.77 | temperature | 25 | 270 | > th | dose independent | rect. | 18 | 22 |
| 703 | 517.05 | temperature | 25 | 90 | < th | dose independent | rect. | 11 | 23 |
| 746 | 517.41 | temperature | 20 | 10 | > th | dose dependent | rect. | 21 | 19 |
| 772 | 517.64 | temperature | 25 | 270 | > th | dose independent | rect. | 13 | 23 |
| 780 | 517.68 | humidity | 50 | 43200 | < th | dose independent | rect. | 16 | 18 |
| 816 | 517.98 | humidity | 80 | 30 | < th | dose independent | rect. | 8 | 10 |
| 885 | 518.48 | temperature | 25 | 10 | > th | dose independent | rect. | 19 | 17 |
| 889 | 518.52 | temperature | 25 | 10 | > th | dose independent | rect. | 21 | 23 |
| 920 | 518.77 | temperature | 25 | 10 | > th | dose independent | rect. | 1 | 23 |
| 929 | 518.84 | temperature | 30 | 10 | < th | dose dependent | no | NA | NA |
| 942 | 518.97 | temperature | 25 | 10 | > th | dose independent | rect. | 4 | 20 |
| 969 | 519.13 | temperature | 20 | 720 | > th | dose dependent | rect. | 21 | 9 |
| 979 | 519.23 | temperature | 30 | 10 | < th | dose dependent | rect. | 5 | 23 |
| 984 | 519.30 | temperature | 25 | 270 | < th | dose independent | rect. | 9 | 23 |
| 1000 | 519.39 | humidity | 50 | 43200 | < th | dose dependent | rect. | 16 | 1 |