Os04g0494800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF642 family protein.
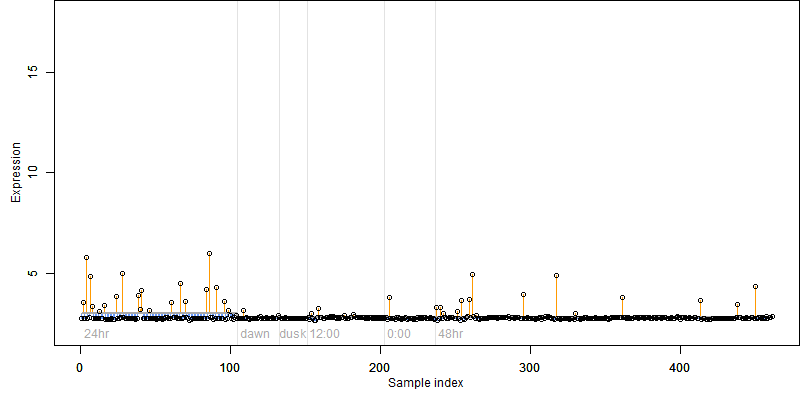

FiT-DB / Search/ Help/ Sample detail
|
Os04g0494800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF642 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
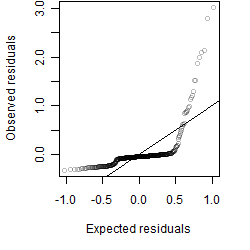
Dependence on each variable

Residual plot

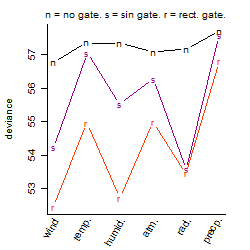
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 56.7 | 0.354 | 2.81 | -0.166 | 0.0611 | -0.184 | -0.00609 | 1.07 | 0.157 | 9.57 | 1 | 15 | -- | -- |
| 57.7 | 0.354 | 2.8 | -0.129 | 0.0639 | -0.102 | 0.0314 | -- | 0.171 | 9.77 | 1.212 | 15 | -- | -- |
| 56.7 | 0.351 | 2.81 | -0.166 | 0.061 | -0.185 | -- | 1.07 | 0.157 | 9.64 | 1 | 14 | -- | -- |
| 57.63 | 0.356 | 2.8 | -0.143 | 0.0651 | -0.0798 | -- | -- | 0.168 | 10.6 | 2.053 | 12 | -- | -- |
| 56.86 | 0.351 | 2.81 | -0.185 | -- | -0.167 | -- | 0.964 | 0.158 | -- | 1.231 | 16 | -- | -- |
| 57.94 | 0.357 | 2.8 | -0.109 | 0.0807 | -- | -0.203 | -- | 0.165 | 6.46 | -- | -- | -- | -- |
| 57.99 | 0.357 | 2.8 | -0.111 | 0.0837 | -- | -- | -- | 0.167 | 7.72 | -- | -- | -- | -- |
| 57.87 | 0.354 | 2.8 | -0.145 | -- | -0.0843 | -- | -- | 0.171 | -- | 2.067 | 12 | -- | -- |
| 58.13 | 0.357 | 2.79 | -- | 0.0663 | -0.0555 | -- | -- | 0.182 | 9.77 | 2 | 12 | -- | -- |
| 58.37 | 0.357 | 2.8 | -0.11 | -- | -- | -- | -- | 0.168 | -- | -- | -- | -- | -- |
| 58.32 | 0.357 | 2.79 | -- | 0.0829 | -- | -- | -- | 0.179 | 7.81 | -- | -- | -- | -- |
| 58.39 | 0.357 | 2.79 | -- | -- | -0.0575 | -- | -- | 0.182 | -- | 2.067 | 12 | -- | -- |
| 58.69 | 0.358 | 2.8 | -- | -- | -- | -- | -- | 0.18 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram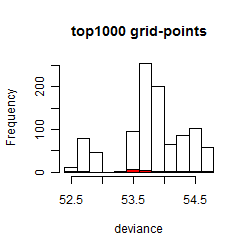 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 52.45 | wind | 1 | 10 | < th | dose independent | rect. | 6 | 4 |
| 13 | 52.72 | humidity | 60 | 10 | < th | dose independent | rect. | 10 | 1 |
| 15 | 52.72 | humidity | 60 | 10 | < th | dose dependent | rect. | 10 | 1 |
| 141 | 53.44 | humidity | 60 | 270 | < th | dose dependent | rect. | 19 | 14 |
| 142 | 53.45 | radiation | 40 | 10 | > th | dose dependent | rect. | 9 | 1 |
| 184 | 53.52 | wind | 1 | 10 | < th | dose dependent | rect. | 6 | 6 |
| 186 | 53.53 | wind | 1 | 10 | < th | dose dependent | rect. | 6 | 4 |
| 191 | 53.57 | radiation | 40 | 10 | > th | dose independent | rect. | 9 | 1 |
| 211 | 53.57 | radiation | 40 | 10 | > th | dose dependent | sin | 12 | NA |
| 259 | 53.66 | humidity | 60 | 1440 | > th | dose independent | rect. | 8 | 1 |
| 261 | 53.67 | humidity | 60 | 1440 | > th | dose independent | rect. | 21 | 12 |
| 384 | 53.71 | wind | 1 | 10 | < th | dose dependent | rect. | 9 | 1 |
| 442 | 53.76 | wind | 1 | 10 | < th | dose independent | rect. | 9 | 1 |
| 478 | 53.77 | humidity | 60 | 1440 | > th | dose independent | rect. | 24 | 9 |
| 682 | 53.99 | humidity | 60 | 1440 | > th | dose independent | rect. | 4 | 5 |
| 694 | 54.08 | radiation | 40 | 10 | > th | dose independent | sin | 11 | NA |
| 775 | 54.23 | wind | 1 | 10 | < th | dose independent | sin | 21 | NA |
| 846 | 54.43 | wind | 1 | 10 | > th | dose independent | sin | 22 | NA |
| 967 | 54.68 | wind | 1 | 10 | < th | dose dependent | sin | 21 | NA |