Os04g0376100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Mitochondrial import inner membrane translocase, subunit Tim17/22 family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os04g0376100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Mitochondrial import inner membrane translocase, subunit Tim17/22 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16.37 | 0.19 | 11.4 | -0.133 | 0.284 | -0.759 | -0.209 | 0.703 | -0.0261 | 20.1 | 24.31 | 1243 | -- | -- |
| 16.76 | 0.191 | 11.3 | -0.0344 | 0.29 | -0.914 | -0.255 | -- | 0.00489 | 20.3 | 23.3 | 1306 | -- | -- |
| 16.47 | 0.189 | 11.4 | -0.124 | 0.271 | -0.746 | -- | 0.711 | -0.0235 | 19.9 | 24.29 | 1141 | -- | -- |
| 16.92 | 0.192 | 11.3 | -0.0144 | 0.28 | -0.888 | -- | -- | 0.00912 | 20 | 23.38 | 1213 | -- | -- |
| 17.83 | 0.197 | 11.4 | -0.274 | -- | -0.867 | -- | 1.68 | -0.0483 | -- | 33.96 | 686 | -- | -- |
| 26.19 | 0.24 | 11.3 | 0.229 | 0.292 | -- | -0.222 | -- | 0.162 | 20.8 | -- | -- | -- | -- |
| 26.36 | 0.24 | 11.3 | 0.237 | 0.294 | -- | -- | -- | 0.162 | 20.7 | -- | -- | -- | -- |
| 19.52 | 0.206 | 11.3 | -0.000264 | -- | -0.835 | -- | -- | -0.0253 | -- | 26.62 | 692 | -- | -- |
| 16.92 | 0.192 | 11.3 | -- | 0.279 | -0.88 | -- | -- | 0.0107 | 20 | 23.4 | 1213 | -- | -- |
| 31.24 | 0.261 | 11.3 | 0.236 | -- | -- | -- | -- | 0.16 | -- | -- | -- | -- | -- |
| 27.83 | 0.247 | 11.3 | -- | 0.293 | -- | -- | -- | 0.136 | 20.8 | -- | -- | -- | -- |
| 19.52 | 0.206 | 11.3 | -- | -- | -0.835 | -- | -- | -0.0252 | -- | 26.62 | 692 | -- | -- |
| 32.71 | 0.267 | 11.3 | -- | -- | -- | -- | -- | 0.134 | -- | -- | -- | -- | -- |
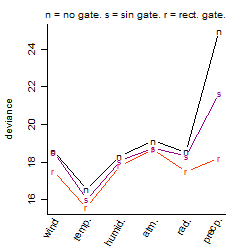
Results of the grid search
Summarized heatmap of deviance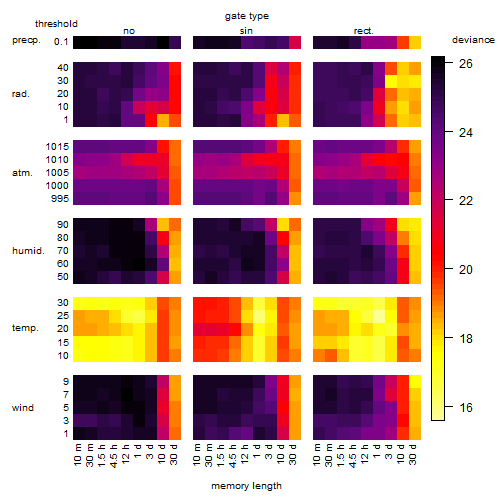
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 15.59 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 1 |
| 20 | 15.88 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 12 |
| 44 | 16.01 | temperature | 25 | 1440 | < th | dose dependent | sin | 13 | NA |
| 190 | 16.48 | temperature | 25 | 1440 | < th | dose dependent | rect. | 4 | 1 |
| 207 | 16.52 | temperature | 25 | 1440 | < th | dose dependent | no | NA | NA |
| 214 | 16.53 | temperature | 10 | 1440 | > th | dose dependent | rect. | 20 | 10 |
| 223 | 16.54 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 23 |
| 256 | 16.59 | temperature | 10 | 1440 | > th | dose dependent | rect. | 4 | 1 |
| 258 | 16.59 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 5 |
| 309 | 16.64 | temperature | 10 | 1440 | > th | dose dependent | rect. | 21 | 3 |
| 414 | 16.72 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 9 |
| 490 | 16.77 | temperature | 10 | 270 | > th | dose dependent | rect. | 3 | 23 |
| 525 | 16.80 | temperature | 10 | 1440 | > th | dose dependent | sin | 11 | NA |
| 535 | 16.80 | temperature | 30 | 270 | < th | dose dependent | rect. | 7 | 23 |
| 561 | 16.83 | temperature | 20 | 4320 | < th | dose independent | rect. | 19 | 4 |
| 593 | 16.85 | temperature | 20 | 4320 | > th | dose independent | rect. | 18 | 5 |
| 677 | 16.90 | temperature | 30 | 90 | < th | dose dependent | rect. | 9 | 23 |
| 688 | 16.91 | temperature | 15 | 270 | > th | dose dependent | rect. | 15 | 23 |
| 765 | 16.98 | temperature | 20 | 4320 | < th | dose independent | rect. | 21 | 1 |
| 807 | 17.01 | temperature | 20 | 4320 | > th | dose independent | rect. | 21 | 1 |
| 847 | 17.03 | temperature | 10 | 720 | > th | dose dependent | rect. | 11 | 23 |
| 867 | 17.05 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 23 |
| 954 | 17.12 | temperature | 30 | 10 | < th | dose dependent | rect. | 11 | 23 |
| 971 | 17.14 | temperature | 15 | 720 | > th | dose dependent | no | NA | NA |
| 986 | 17.14 | temperature | 15 | 720 | > th | dose dependent | rect. | 8 | 23 |