Os04g0354600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Acyl CoA reductase-like protein.

FiT-DB / Search/ Help/ Sample detail
|
Os04g0354600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Acyl CoA reductase-like protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
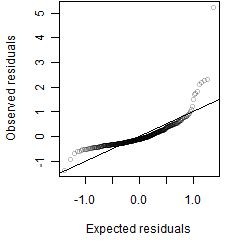
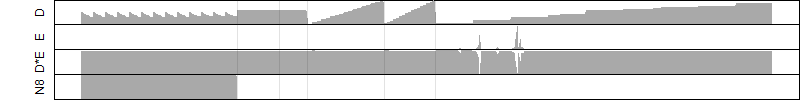
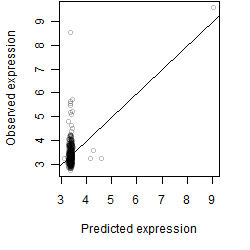 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose independent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 104.1 | 0.479 | 3.24 | -0.344 | 0.281 | -16.2 | -0.236 | -74.8 | 0.00297 | 6.06 | 9.016 | 88 | 6.83 | -- |
| 141.16 | 0.558 | 3.38 | 0.0188 | 0.181 | -0.344 | 0.53 | -- | -0.0358 | 7.03 | 9.016 | 88 | 6.83 | -- |
| 104.2 | 0.475 | 3.24 | -0.348 | 0.278 | -16.2 | -- | -74.8 | 0.00224 | 6.29 | 9.012 | 86 | 6.69 | -- |
| 133.77 | 0.539 | 3.38 | 0.0589 | 0.229 | 1.64 | -- | -- | -0.0146 | 6.4 | 9.077 | 44 | 6.21 | -- |
| 108.14 | 0.484 | 3.25 | -0.345 | -- | -16.2 | -- | -73.6 | 0.00402 | -- | 9 | 86 | 6.65 | -- |
| 141.27 | 0.557 | 3.38 | 0.0224 | 0.184 | -- | 0.52 | -- | -0.0345 | 7.03 | -- | -- | -- | -- |
| 141.94 | 0.558 | 3.38 | 0.0313 | 0.189 | -- | -- | -- | -0.0363 | 6.48 | -- | -- | -- | -- |
| 136.16 | 0.543 | 3.38 | 0.131 | -- | 1.62 | -- | -- | 0.0212 | -- | 9 | 44 | 7.98 | -- |
| 133.23 | 0.538 | 3.38 | -- | 0.251 | 1.76 | -- | -- | -0.0125 | 6.33 | 9.062 | 44 | 8.08 | -- |
| 143.79 | 0.56 | 3.39 | 0.036 | -- | -- | -- | -- | -0.0324 | -- | -- | -- | -- | -- |
| 141.96 | 0.557 | 3.38 | -- | 0.189 | -- | -- | -- | -0.0398 | 6.47 | -- | -- | -- | -- |
| 136.54 | 0.544 | 3.38 | -- | -- | 1.59 | -- | -- | -0.00905 | -- | 9 | 44 | 8.05 | -- |
| 143.82 | 0.56 | 3.39 | -- | -- | -- | -- | -- | -0.0364 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance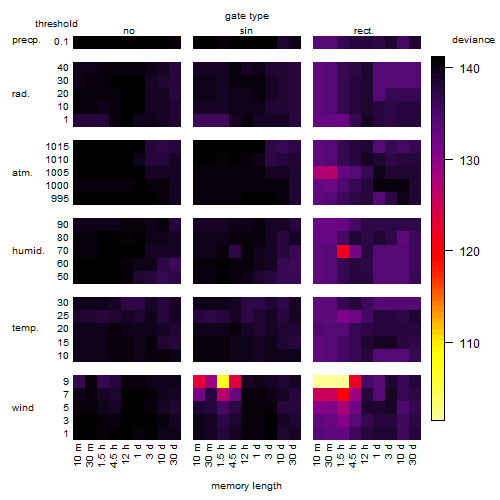
|
Histogram 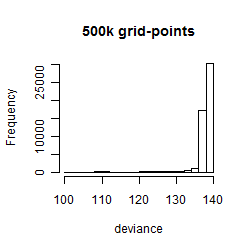
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 101.45 | wind | 9 | 10 | > th | dose independent | rect. | 18 | 1 |
| 4 | 101.45 | wind | 9 | 10 | > th | dose dependent | rect. | 18 | 1 |
| 8 | 103.46 | wind | 9 | 90 | > th | dose dependent | rect. | 17 | 9 |
| 25 | 103.73 | wind | 9 | 90 | > th | dose independent | rect. | 17 | 9 |
| 75 | 106.45 | wind | 9 | 90 | > th | dose independent | sin | 13 | NA |
| 77 | 107.34 | wind | 9 | 90 | > th | dose dependent | sin | 13 | NA |
| 80 | 108.71 | wind | 9 | 90 | < th | dose independent | sin | 12 | NA |
| 82 | 108.74 | wind | 9 | 90 | > th | dose independent | rect. | 13 | 6 |
| 88 | 108.76 | wind | 9 | 90 | > th | dose independent | rect. | 13 | 13 |
| 153 | 110.67 | wind | 9 | 90 | > th | dose dependent | rect. | 13 | 6 |
| 159 | 110.68 | wind | 9 | 90 | > th | dose dependent | rect. | 13 | 13 |
| 303 | 121.69 | humidity | 70 | 90 | < th | dose independent | rect. | 16 | 1 |
| 341 | 123.06 | wind | 9 | 10 | > th | dose independent | sin | 12 | NA |
| 527 | 125.51 | wind | 9 | 10 | < th | dose independent | sin | 12 | NA |
| 558 | 126.30 | wind | 7 | 10 | > th | dose dependent | rect. | 17 | 12 |
| 573 | 126.40 | atmosphere | 1005 | 30 | > th | dose dependent | rect. | 17 | 1 |
| 608 | 126.93 | wind | 9 | 10 | < th | dose independent | sin | 14 | NA |
| 610 | 126.97 | wind | 9 | 270 | > th | dose independent | rect. | 13 | 1 |
| 614 | 126.97 | wind | 9 | 270 | > th | dose dependent | rect. | 13 | 1 |
| 796 | 129.34 | atmosphere | 1005 | 30 | > th | dose independent | rect. | 17 | 1 |
| 842 | 130.33 | wind | 9 | 10 | < th | dose independent | rect. | 5 | 23 |
| 907 | 131.27 | temperature | 25 | 90 | < th | dose independent | rect. | 16 | 1 |
| 908 | 131.28 | temperature | 25 | 90 | > th | dose dependent | rect. | 2 | 3 |
| 944 | 131.55 | wind | 7 | 10 | > th | dose dependent | sin | 11 | NA |
| 946 | 131.58 | temperature | 25 | 90 | > th | dose independent | rect. | 1 | 4 |
| 965 | 131.99 | temperature | 25 | 270 | > th | dose dependent | rect. | 2 | 1 |
| 966 | 131.99 | radiation | 1 | 90 | < th | dose dependent | rect. | 17 | 1 |
| 970 | 132.05 | humidity | 90 | 10 | < th | dose dependent | rect. | 4 | 1 |
| 983 | 132.17 | temperature | 25 | 90 | < th | dose independent | rect. | 16 | 3 |
| 990 | 132.26 | atmosphere | 1005 | 30 | > th | dose dependent | rect. | 2 | 3 |