Os04g0228400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Expansin precursor (Alpha-expansin OsEXPA1).

FiT-DB / Search/ Help/ Sample detail
|
Os04g0228400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Expansin precursor (Alpha-expansin OsEXPA1).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
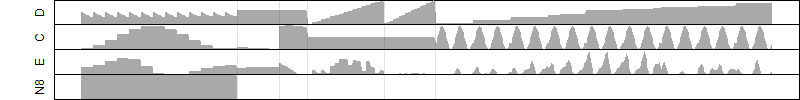
 __
__
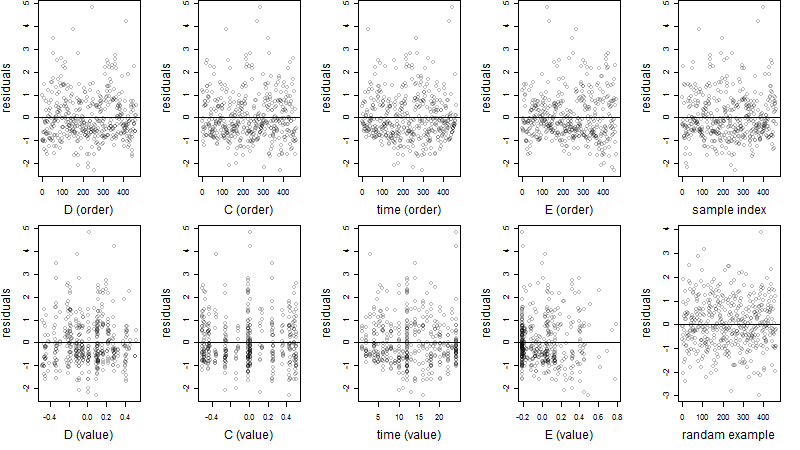
Dependence on each variable

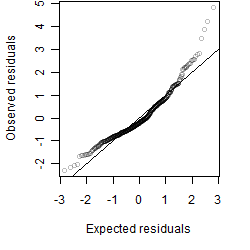
Residual plot
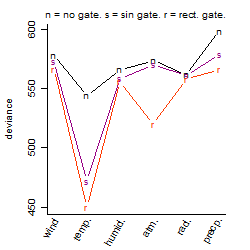
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 445.71 | 0.992 | 4.22 | -1.16 | 1.56 | 4.27 | -1.37 | -1.68 | -0.641 | 18.1 | 21.01 | 277 | 23.8 | 14.8 |
| 447.3 | 0.985 | 4.21 | -0.991 | 1.59 | 4.2 | -1.65 | -- | -0.595 | 17.9 | 21.06 | 276 | 23.8 | 14.8 |
| 449.66 | 0.988 | 4.23 | -1.17 | 1.59 | 4.3 | -- | -1.83 | -0.648 | 18.1 | 21.09 | 281 | 23.7 | 14.8 |
| 451.66 | 0.99 | 4.19 | -0.943 | 1.63 | 4.25 | -- | -- | -0.561 | 18.1 | 21.04 | 284 | 23.8 | 14.5 |
| 513.08 | 1.05 | 4.21 | -1.15 | -- | 4.55 | -- | -1.75 | -0.612 | -- | 22.17 | 290 | 0.867 | 14.8 |
| 671.55 | 1.21 | 4.05 | -0.311 | 2.2 | -- | -0.546 | -- | -0.0501 | 15 | -- | -- | -- | -- |
| 672.52 | 1.21 | 4.05 | -0.323 | 2.2 | -- | -- | -- | -0.0534 | 15 | -- | -- | -- | -- |
| 488.88 | 1.03 | 4.09 | -0.894 | -- | 4.26 | -- | -- | -0.378 | -- | 21.01 | 296 | 7.52 | 7.97 |
| 474.33 | 1.01 | 4.18 | -- | 1.59 | 4.11 | -- | -- | -0.478 | 17.8 | 21.3 | 281 | 23.9 | 14.5 |
| 950.15 | 1.44 | 4.06 | -0.417 | -- | -- | -- | -- | -0.111 | -- | -- | -- | -- | -- |
| 675.27 | 1.22 | 4.05 | -- | 2.21 | -- | -- | -- | -0.0174 | 15 | -- | -- | -- | -- |
| 498.98 | 1.04 | 4.08 | -- | -- | 4.53 | -- | -- | -0.197 | -- | 19.24 | 367 | 7.84 | 6.47 |
| 954.74 | 1.44 | 4.05 | -- | -- | -- | -- | -- | -0.0643 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 449.85 | temperature | 20 | 270 | > th | dose dependent | rect. | 24 | 15 |
| 28 | 471.99 | temperature | 20 | 270 | > th | dose dependent | sin | 4 | NA |
| 31 | 474.77 | temperature | 30 | 270 | < th | dose dependent | rect. | 5 | 14 |
| 33 | 475.63 | temperature | 20 | 270 | > th | dose dependent | rect. | 8 | 7 |
| 49 | 481.17 | temperature | 30 | 270 | < th | dose dependent | rect. | 4 | 7 |
| 54 | 482.73 | temperature | 30 | 270 | < th | dose dependent | sin | 2 | NA |
| 95 | 490.85 | temperature | 25 | 270 | > th | dose independent | rect. | 8 | 8 |
| 171 | 500.91 | temperature | 25 | 270 | < th | dose independent | rect. | 5 | 14 |
| 189 | 503.02 | temperature | 25 | 270 | < th | dose independent | rect. | 5 | 6 |
| 218 | 506.12 | temperature | 25 | 270 | > th | dose independent | rect. | 24 | 15 |
| 229 | 507.12 | temperature | 20 | 10 | > th | dose dependent | rect. | 2 | 15 |
| 261 | 509.28 | temperature | 25 | 270 | > th | dose independent | sin | 1 | NA |
| 330 | 514.94 | temperature | 10 | 270 | > th | dose dependent | rect. | 8 | 22 |
| 333 | 515.55 | temperature | 25 | 270 | > th | dose dependent | rect. | 11 | 1 |
| 350 | 516.31 | temperature | 25 | 270 | < th | dose independent | sin | 1 | NA |
| 409 | 519.88 | atmosphere | 1015 | 270 | < th | dose dependent | rect. | 9 | 6 |
| 411 | 519.91 | temperature | 25 | 10 | > th | dose dependent | rect. | 11 | 6 |
| 545 | 526.00 | temperature | 25 | 10 | > th | dose dependent | rect. | 9 | 8 |
| 598 | 527.62 | temperature | 30 | 720 | < th | dose dependent | rect. | 22 | 14 |
| 694 | 531.32 | temperature | 30 | 10 | < th | dose dependent | rect. | 6 | 7 |
| 746 | 532.74 | temperature | 30 | 10 | < th | dose dependent | rect. | 7 | 15 |
| 811 | 534.23 | temperature | 30 | 270 | < th | dose dependent | rect. | 14 | 22 |
| 852 | 535.00 | atmosphere | 995 | 270 | > th | dose dependent | rect. | 5 | 15 |
| 926 | 536.92 | atmosphere | 1010 | 270 | < th | dose independent | rect. | 9 | 7 |