Os04g0110200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.


FiT-DB / Search/ Help/ Sample detail
|
Os04g0110200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

Dependence on each variable

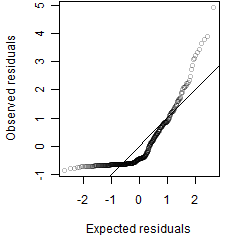
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 356.38 | 0.887 | 3.49 | -1.33 | 0.127 | -0.449 | 0.82 | 2.03 | -0.631 | 0.62 | 24.7 | 270 | -- | -- |
| 367.25 | 0.893 | 3.44 | -0.784 | 0.0701 | -0.47 | 1.26 | -- | -0.515 | 1.07 | 24.1 | 254 | -- | -- |
| 359.11 | 0.883 | 3.5 | -1.43 | 0.121 | -0.45 | -- | 2.24 | -0.652 | 0.661 | 24.7 | 269 | -- | -- |
| 372.34 | 0.899 | 3.43 | -0.742 | 0.116 | -0.365 | -- | -- | -0.406 | 23.2 | 24.1 | 283 | -- | -- |
| 359.56 | 0.883 | 3.5 | -1.42 | -- | -0.443 | -- | 2.13 | -0.651 | -- | 24.5 | 271 | -- | -- |
| 375.81 | 0.909 | 3.38 | -0.585 | 0.0983 | -- | -1.48 | -- | -0.236 | 15.3 | -- | -- | -- | -- |
| 381 | 0.914 | 3.39 | -0.614 | 0.209 | -- | -- | -- | -0.243 | 19.2 | -- | -- | -- | -- |
| 373 | 0.9 | 3.42 | -0.762 | -- | -0.386 | -- | -- | -0.414 | -- | 24.2 | 278 | -- | -- |
| 386.82 | 0.916 | 3.4 | -- | 0.166 | -0.278 | -- | -- | -0.265 | 21.9 | 24.2 | 278 | -- | -- |
| 383.32 | 0.915 | 3.39 | -0.618 | -- | -- | -- | -- | -0.247 | -- | -- | -- | -- | -- |
| 390.93 | 0.925 | 3.37 | -- | 0.216 | -- | -- | -- | -0.175 | 19 | -- | -- | -- | -- |
| 387.62 | 0.917 | 3.4 | -- | -- | -0.28 | -- | -- | -0.291 | -- | 24.1 | 278 | -- | -- |
| 393.38 | 0.926 | 3.37 | -- | -- | -- | -- | -- | -0.178 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram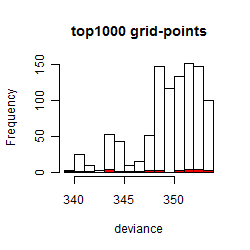 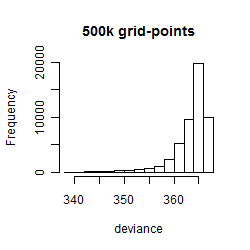
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 339.37 | humidity | 80 | 30 | < th | dose dependent | rect. | 15 | 18 |
| 3 | 340.06 | humidity | 80 | 10 | < th | dose dependent | rect. | 15 | 6 |
| 30 | 341.62 | humidity | 70 | 270 | < th | dose dependent | rect. | 15 | 1 |
| 39 | 343.10 | radiation | 40 | 90 | > th | dose independent | rect. | 15 | 1 |
| 56 | 343.10 | radiation | 40 | 90 | > th | dose dependent | rect. | 15 | 1 |
| 83 | 343.35 | humidity | 80 | 10 | < th | dose dependent | sin | 13 | NA |
| 90 | 344.00 | humidity | 60 | 270 | < th | dose independent | rect. | 15 | 1 |
| 118 | 344.82 | humidity | 70 | 10 | < th | dose independent | rect. | 15 | 5 |
| 139 | 345.68 | atmosphere | 1005 | 270 | < th | dose dependent | rect. | 10 | 7 |
| 147 | 346.42 | humidity | 70 | 90 | < th | dose independent | rect. | 15 | 16 |
| 161 | 347.31 | humidity | 70 | 90 | < th | dose independent | rect. | 15 | 5 |
| 196 | 347.82 | humidity | 80 | 270 | < th | dose dependent | rect. | 12 | 8 |
| 215 | 348.10 | humidity | 80 | 270 | < th | dose dependent | rect. | 12 | 20 |
| 328 | 348.82 | humidity | 60 | 270 | < th | dose independent | rect. | 13 | 6 |
| 494 | 350.22 | humidity | 70 | 90 | < th | dose independent | sin | 13 | NA |
| 590 | 350.86 | humidity | 50 | 10 | > th | dose dependent | sin | 15 | NA |
| 599 | 350.93 | humidity | 60 | 270 | > th | dose independent | sin | 14 | NA |
| 617 | 351.13 | humidity | 80 | 30 | < th | dose dependent | rect. | 15 | 1 |
| 677 | 351.47 | humidity | 60 | 10 | > th | dose dependent | rect. | 12 | 14 |
| 707 | 351.58 | humidity | 70 | 10 | > th | dose dependent | rect. | 9 | 17 |
| 715 | 351.68 | humidity | 50 | 270 | > th | dose dependent | sin | 17 | NA |
| 765 | 352.08 | atmosphere | 1005 | 720 | < th | dose independent | rect. | 9 | 5 |
| 779 | 352.14 | humidity | 70 | 10 | < th | dose independent | sin | 13 | NA |
| 793 | 352.24 | temperature | 25 | 270 | > th | dose independent | rect. | 13 | 20 |
| 858 | 352.75 | radiation | 10 | 1440 | > th | dose independent | rect. | 9 | 5 |
| 930 | 353.09 | wind | 5 | 270 | < th | dose independent | rect. | 15 | 1 |
| 945 | 353.10 | atmosphere | 1005 | 270 | < th | dose dependent | sin | 21 | NA |
| 946 | 353.10 | humidity | 60 | 10 | > th | dose dependent | rect. | 14 | 12 |