Os04g0103400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Non-protein coding transcript, uncharacterized transcript.


FiT-DB / Search/ Help/ Sample detail
|
Os04g0103400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Non-protein coding transcript, uncharacterized transcript.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
Dependence on each variable

Residual plot

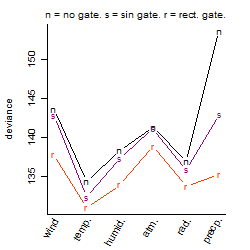
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 133.18 | 0.542 | 5.48 | -2.17 | 1.18 | 1.7 | -0.96 | -0.607 | -0.273 | 16.2 | 17.66 | 4540 | -- | -- |
| 133.04 | 0.537 | 5.47 | -2.1 | 1.19 | 1.74 | -0.948 | -- | -0.292 | 16.2 | 16.42 | 5879 | -- | -- |
| 135.75 | 0.543 | 5.48 | -2.18 | 1.17 | 1.69 | -- | -0.599 | -0.276 | 16.2 | 17.85 | 4539 | -- | -- |
| 135.25 | 0.542 | 5.47 | -2.11 | 1.18 | 1.73 | -- | -- | -0.305 | 16.3 | 16.6 | 5878 | -- | -- |
| 177.58 | 0.624 | 5.47 | -1.9 | -- | 2.17 | -- | -1.4 | -0.296 | -- | 17.85 | 4539 | -- | -- |
| 198.24 | 0.66 | 5.37 | -1.19 | 1.14 | -- | -0.973 | -- | 0.133 | 16.3 | -- | -- | -- | -- |
| 200.84 | 0.664 | 5.37 | -1.2 | 1.13 | -- | -- | -- | 0.128 | 16.3 | -- | -- | -- | -- |
| 179.66 | 0.627 | 5.44 | -1.69 | -- | 2.26 | -- | -- | -0.234 | -- | 16.6 | 5878 | -- | -- |
| 214.64 | 0.682 | 5.4 | -- | 1.09 | 0.961 | -- | -- | 0.123 | 16 | 22.2 | 3609 | -- | -- |
| 270.12 | 0.768 | 5.37 | -1.25 | -- | -- | -- | -- | 0.0978 | -- | -- | -- | -- | -- |
| 239 | 0.723 | 5.34 | -- | 1.16 | -- | -- | -- | 0.262 | 16.3 | -- | -- | -- | -- |
| 250.85 | 0.74 | 5.39 | -- | -- | 1.79 | -- | -- | 0.0115 | -- | 16.6 | 5878 | -- | -- |
| 311.21 | 0.823 | 5.34 | -- | -- | -- | -- | -- | 0.236 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 131.01 | temperature | 25 | 270 | < th | dose dependent | rect. | 23 | 21 |
| 3 | 131.59 | temperature | 20 | 14400 | < th | dose dependent | rect. | 4 | 1 |
| 4 | 131.64 | temperature | 20 | 14400 | < th | dose dependent | rect. | 3 | 3 |
| 6 | 131.72 | temperature | 10 | 4320 | > th | dose dependent | rect. | 21 | 3 |
| 8 | 131.73 | temperature | 25 | 4320 | < th | dose dependent | rect. | 21 | 4 |
| 13 | 131.79 | temperature | 25 | 4320 | < th | dose dependent | rect. | 20 | 11 |
| 14 | 131.80 | temperature | 25 | 4320 | < th | dose dependent | rect. | 21 | 7 |
| 15 | 131.80 | temperature | 15 | 4320 | > th | dose dependent | rect. | 20 | 11 |
| 174 | 132.33 | temperature | 15 | 4320 | > th | dose dependent | sin | 11 | NA |
| 175 | 132.33 | temperature | 30 | 4320 | < th | dose dependent | rect. | 6 | 1 |
| 180 | 132.34 | temperature | 30 | 4320 | < th | dose dependent | sin | 11 | NA |
| 198 | 132.40 | temperature | 15 | 4320 | > th | dose dependent | rect. | 5 | 2 |
| 232 | 132.49 | temperature | 15 | 4320 | > th | dose dependent | rect. | 1 | 7 |
| 322 | 132.73 | temperature | 30 | 4320 | < th | dose dependent | rect. | 3 | 5 |
| 503 | 133.17 | temperature | 15 | 14400 | > th | dose dependent | rect. | 3 | 1 |
| 799 | 133.74 | radiation | 1 | 43200 | > th | dose independent | rect. | 17 | 2 |
| 950 | 133.90 | humidity | 90 | 43200 | > th | dose dependent | rect. | 21 | 1 |