Os03g0757300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description :


FiT-DB / Search/ Help/ Sample detail
|
Os03g0757300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description :
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = radiation,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 63.02 | 0.373 | 2.86 | -0.0158 | 0.169 | -0.176 | -0.628 | 0.623 | 0.132 | 2.09 | 30 | 9 | -- | -- |
| 63.72 | 0.372 | 2.86 | -0.0266 | 0.171 | -0.179 | -0.118 | -- | 0.132 | 2.01 | 30 | 6 | -- | -- |
| 63.59 | 0.374 | 2.86 | -0.0196 | 0.184 | -0.198 | -- | 0.179 | 0.134 | 1.42 | 30 | 9 | -- | -- |
| 63.73 | 0.372 | 2.86 | -0.0231 | 0.195 | -0.206 | -- | -- | 0.134 | 1.39 | 30 | 6 | -- | -- |
| 64.4 | 0.376 | 2.86 | -0.00822 | -- | -0.0599 | -- | 0.186 | 0.137 | -- | 30 | 9 | -- | -- |
| 64.05 | 0.375 | 2.86 | -0.0102 | 0.0682 | -- | -0.454 | -- | 0.134 | 5.97 | -- | -- | -- | -- |
| 64.55 | 0.376 | 2.85 | -0.0141 | 0.0658 | -- | -- | -- | 0.136 | 5.8 | -- | -- | -- | -- |
| 64.55 | 0.376 | 2.86 | -0.0137 | -- | -0.0579 | -- | -- | 0.137 | -- | 30 | 9 | -- | -- |
| 63.74 | 0.372 | 2.86 | -- | 0.195 | -0.205 | -- | -- | 0.136 | 1.38 | 30 | 6 | -- | -- |
| 64.77 | 0.376 | 2.86 | -0.0121 | -- | -- | -- | -- | 0.137 | -- | -- | -- | -- | -- |
| 64.56 | 0.376 | 2.85 | -- | 0.0656 | -- | -- | -- | 0.137 | 5.81 | -- | -- | -- | -- |
| 64.55 | 0.375 | 2.86 | -- | -- | -0.0577 | -- | -- | 0.139 | -- | 30 | 6 | -- | -- |
| 64.78 | 0.376 | 2.86 | -- | -- | -- | -- | -- | 0.138 | -- | -- | -- | -- | -- |
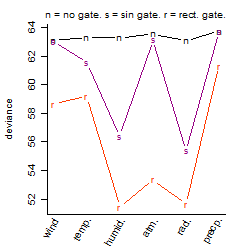
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 51.43 | humidity | 80 | 90 | < th | dose dependent | rect. | 22 | 9 |
| 4 | 51.61 | humidity | 70 | 10 | < th | dose independent | rect. | 18 | 14 |
| 6 | 51.61 | humidity | 70 | 90 | < th | dose dependent | rect. | 19 | 13 |
| 7 | 51.61 | humidity | 70 | 10 | < th | dose dependent | rect. | 18 | 14 |
| 16 | 51.63 | radiation | 30 | 10 | > th | dose independent | rect. | 7 | 1 |
| 32 | 51.63 | humidity | 80 | 90 | < th | dose dependent | rect. | 1 | 6 |
| 67 | 51.64 | radiation | 30 | 10 | > th | dose dependent | rect. | 7 | 1 |
| 73 | 51.64 | humidity | 80 | 90 | < th | dose dependent | rect. | 3 | 4 |
| 105 | 51.65 | humidity | 80 | 90 | < th | dose dependent | rect. | 6 | 1 |
| 126 | 51.66 | humidity | 80 | 90 | < th | dose independent | rect. | 6 | 1 |
| 127 | 51.66 | humidity | 70 | 10 | < th | dose independent | rect. | 7 | 1 |
| 201 | 51.66 | humidity | 70 | 10 | < th | dose dependent | rect. | 7 | 1 |
| 612 | 53.36 | atmosphere | 1005 | 10 | > th | dose dependent | rect. | 7 | 1 |
| 750 | 55.38 | radiation | 30 | 30 | < th | dose independent | sin | 11 | NA |
| 840 | 56.39 | humidity | 70 | 10 | < th | dose independent | sin | 11 | NA |
| 886 | 56.88 | radiation | 20 | 10 | > th | dose independent | sin | 11 | NA |
| 891 | 57.03 | humidity | 70 | 10 | > th | dose independent | sin | 11 | NA |
| 974 | 57.72 | humidity | 70 | 1440 | > th | dose independent | rect. | 1 | 7 |
| 978 | 57.75 | humidity | 80 | 720 | < th | dose dependent | rect. | 21 | 11 |
| 979 | 57.77 | humidity | 70 | 1440 | > th | dose independent | rect. | 7 | 1 |
| 987 | 57.80 | humidity | 70 | 1440 | > th | dose independent | rect. | 22 | 10 |